DataScience_Examples
All about DataSince, DataEngineering and ComputerScience
View the Project on GitHub datainsightat/DataScience_Examples
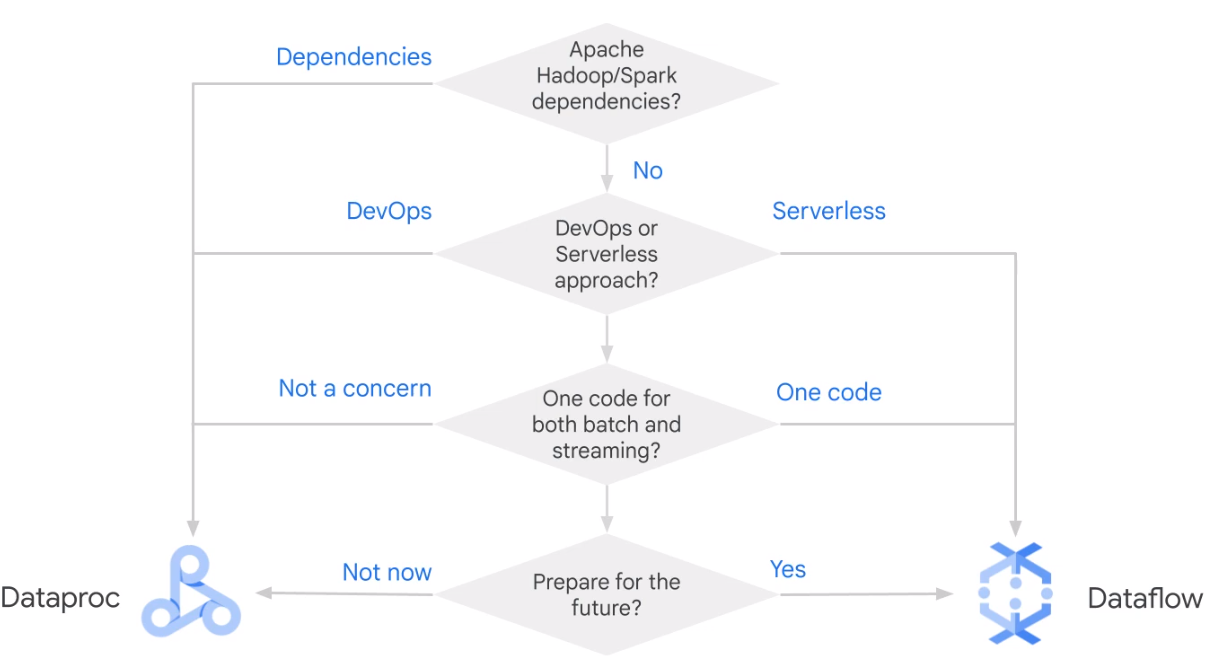
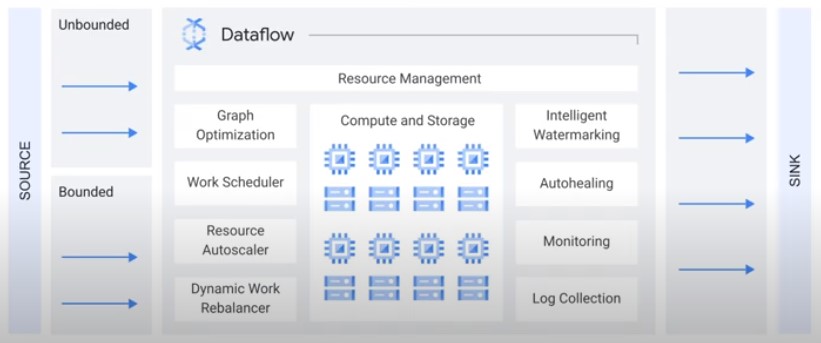
Dataflow
- Scalable
- Low latency
- Batch and Stream
| Dataflow | Dataproc | |
|---|---|---|
| Recommended for | New data processing pipelines, unified batch and streaming | Existing Hadoop/Spak applications, ML, large-batch jobs |
| Fully managed | Yes | No |
| Auto scaling | Yes (adaptive) | Yes (reactive) |
| Expertise | Apache Beam | Hadoop, Hive, Pig, Sprak … |
Apache Beam
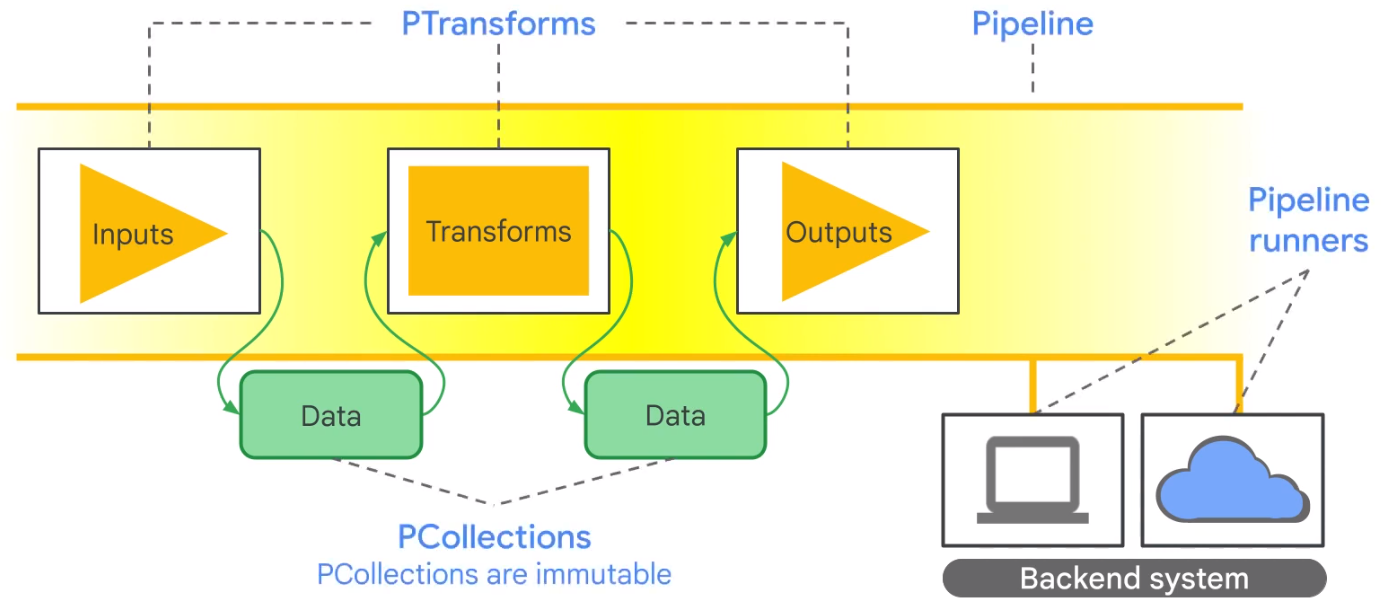
Datagraph
PCollection
In a PCollection all data is immutable and stored as bytestring.
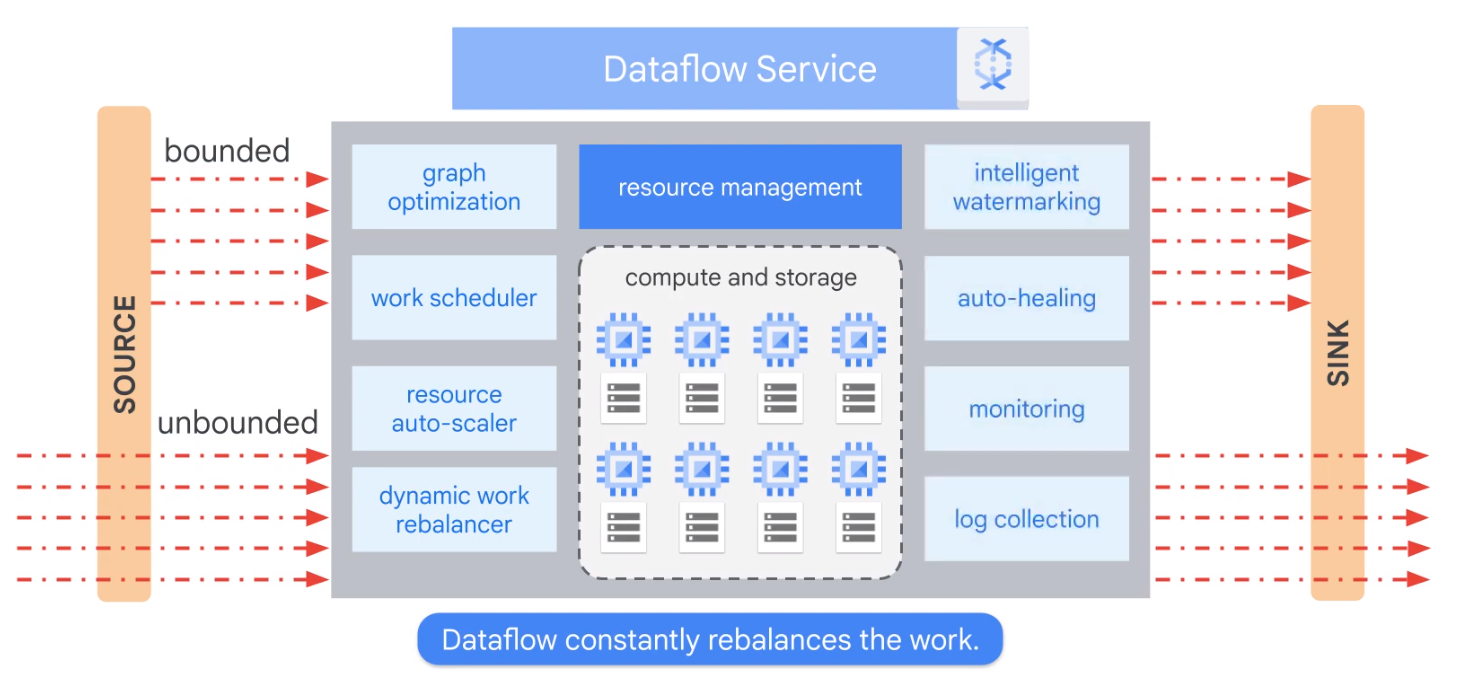
How does Dataflow Work?
- Fully Managed
- Optimizes Datagraph
- Autoscaling
- Rebalancing
- Streaming Semntics
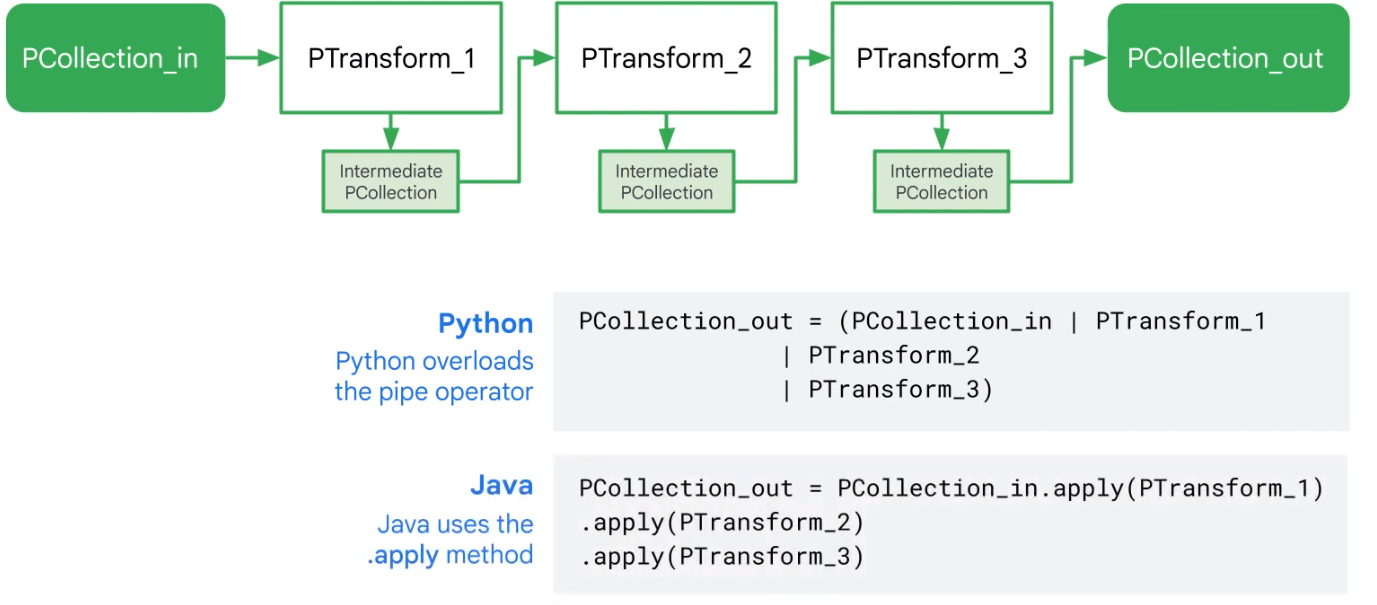
Dataflow Pipelines
Simple Pipeline
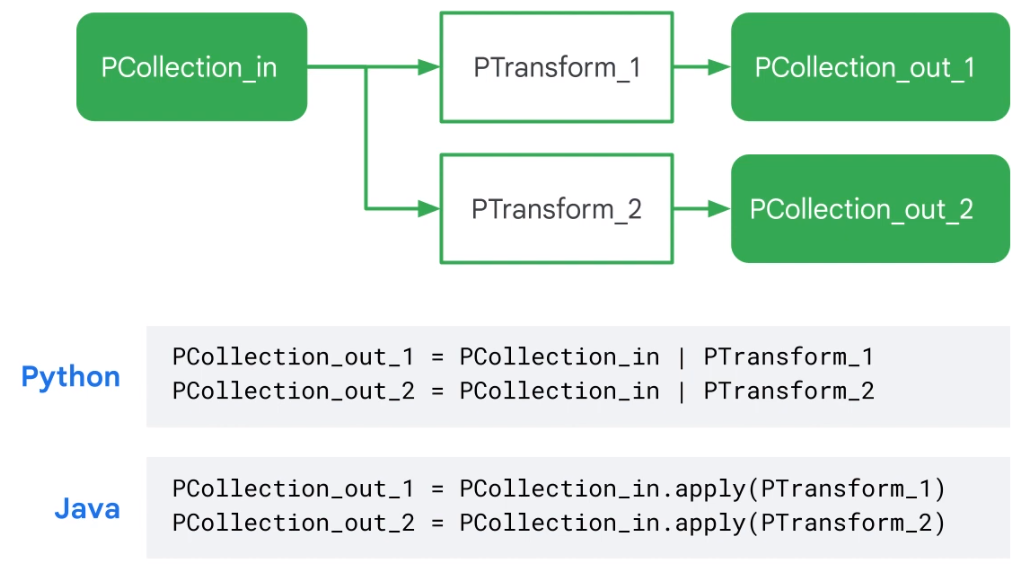
Branching Pipeline
Start End
import apache_beam as beam
if __name == '__main__':
with beam.Pipeline(argv=sys.argv) as p: # Create pipeline parameterized by command line flags
(p
| beam.io.ReadFromText('gs://...') # Read Input
| beam.FlatMap(count_words) # Apply transform
| beam.io.WriteToText('gs://...') # Write output
)
options = {'project':<project>,
'runner':'DataflowRunner'. # Where to run
'region':<region>.
'retup_file':<setup.py file>}
pipeline_options = beam.pipeline.PipelineOptions(flags=[],**options)
pipeline = beam.Pipeline(options=pipeline_options) # Creates the pipeline
Run local
$ python ./grep.py
Run on cloud
$ python ./grep.py \
--project=$PROJECT \
--job_name=myjob \
--staging_location=gs://$BUCKET/staging/ \
--temp_location=gs://$BUCKET/tmp/ \
--runnner)DataflowRunner
Read Data
with beam.Pipeline(options=pipeline_options as p:
# Cloud Storage
lines = p | beam.ioReadFromText("gs://..../input-*.csv.gz")
# Pub/Sub
lines = p | beam.io.ReadStringsFromPubSub(topic=known_args.input_topic)
# BigQuery
query = "select x, y, z from `project.dataset.tablename`"
BQ_source = beam.io.BigQuerySource(query = <query>, use_standard_sql=True)
BQ_data = pipeline | beam.io.Read(BG_srouce)
Write to Sinks
from apache_beam.io.gcp.internal.clients import bigquery
table_spec = bigquery.TableReference(
projectId='clouddataflow-readonly',
datasetId='samples',
tableId='weather_stations')
p | beam.io.WriteToBigQuery(
table_spec,
schema=table_schema,
write_disposition=beam.io.BigQuerryDisposition.WRITE_TRUNCATE,
create_disposition=beam.io.BigQueryDisposition.CREATE_IT_NEEDED)
Map Phase > Transform
'WordLengths' >> beam.Map(word, len(word))
def my_grep(line_term):
if term in line:
yield line
'Grep' >> beam.FlatMap(my_grep(line, searchTerm))
ParDo Parallel Processing
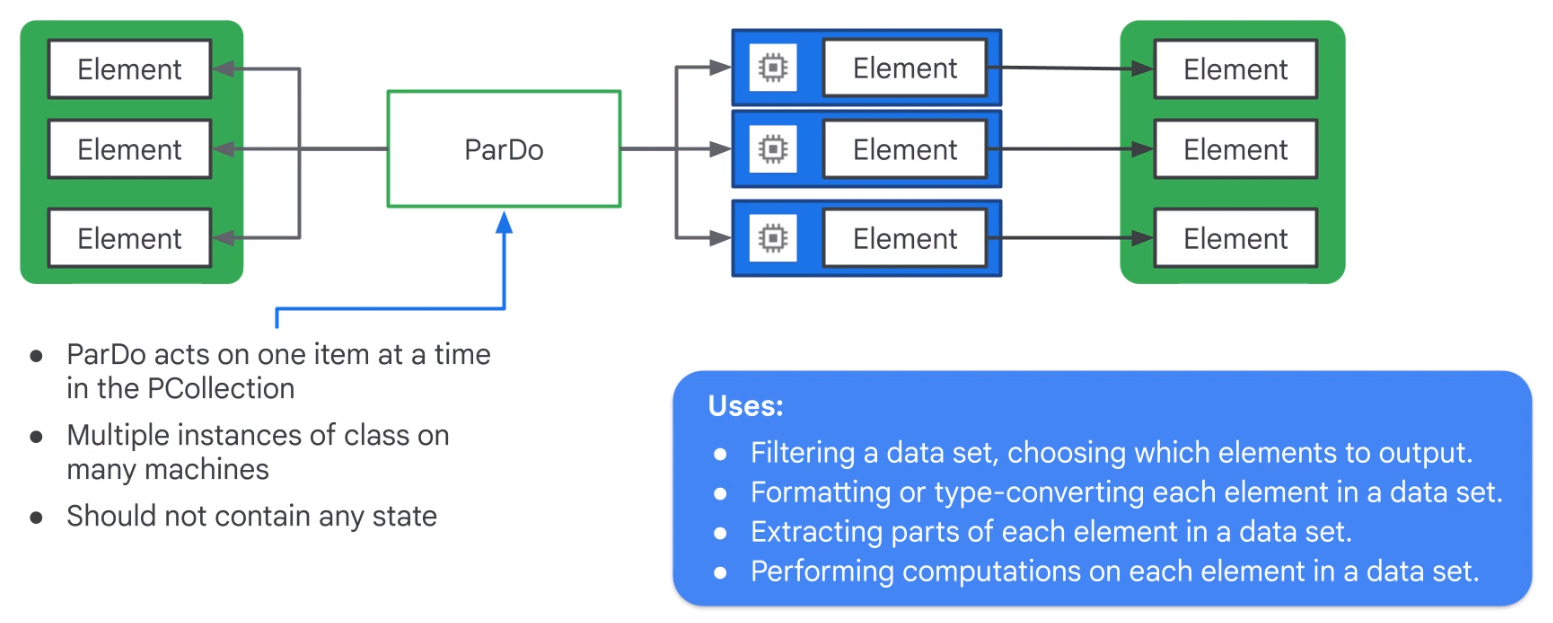
words = ...
# Do Fn
class ComputeWordLengthFn(beam.DoFn):
def process(self,element):
return [len(element)}
# ParDo
word_lengths = words | beam.ParDo(ComputeWordLengthFn())
Multiple Variables
results = (words | beam.PrDo(ProcessWords(),
cutoff_length=2, marker='x')
.with_putputs('above_cutoff_lengths','marked strings',main='below_cutoff_strings'))
below = results.below_cutoff_strings
above = results.above_cutoff_strings
marked = results['marked strings']
GroupByKey
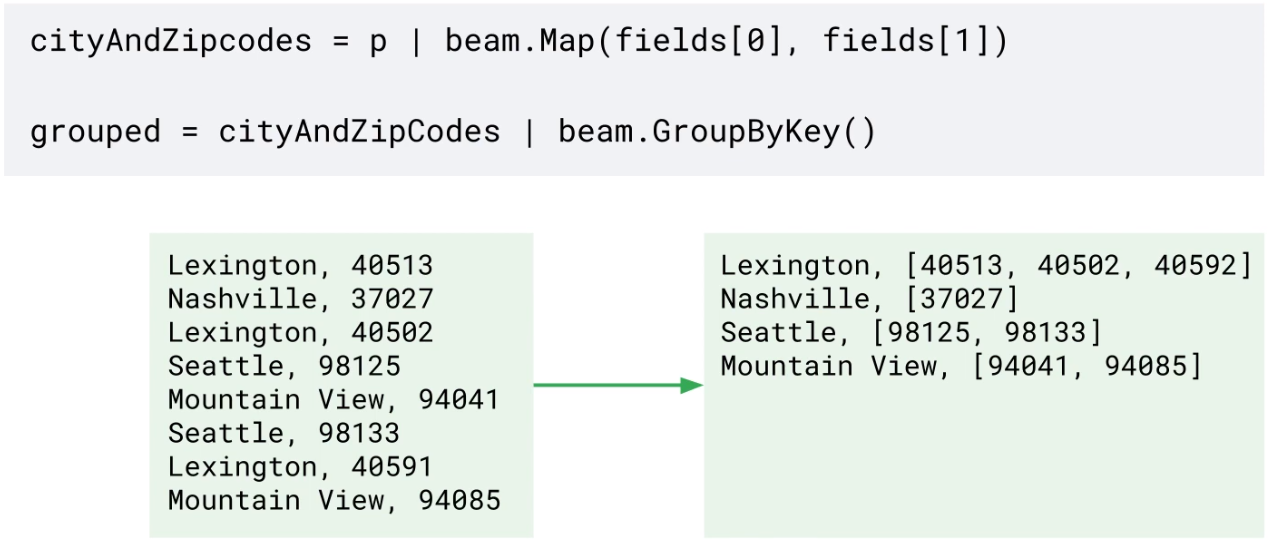
Data skew makes grouping less efficient at scale.
totalAmount = salesAmounts | CombineGlobally(sum)
totalSalesPerPerson = salesRecords | CombinePerKey(sum)
CombineFn works by overriding exisintg operations
You must provide four functions
class AverageFn(beam.CombineFn):
def create_accumulator(self):
return(0.0,0)
def add_input(self, sum_count, input):
(sum, count) = sum_count
return sum + input, count + 1
def merge_accumulators(self, accumulators):
sums, counts = zip(*accumulators)
return sum(sums), sum(counts)
def extract_output(self, sum_count):
(sum, count) = sum_count
return sum / count if count else float('NaN')
pc = ...
average = pc | beam.CombineGlobally(AverageFn())
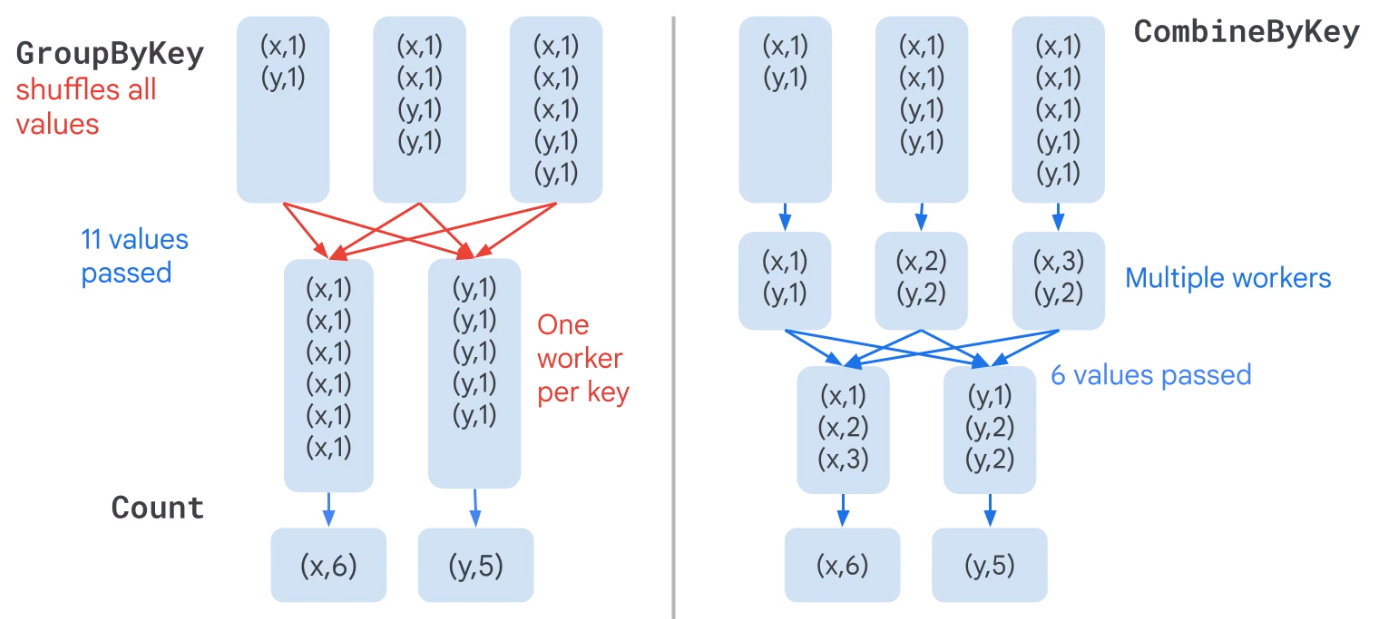
Combine is more efficient that GroupByKey
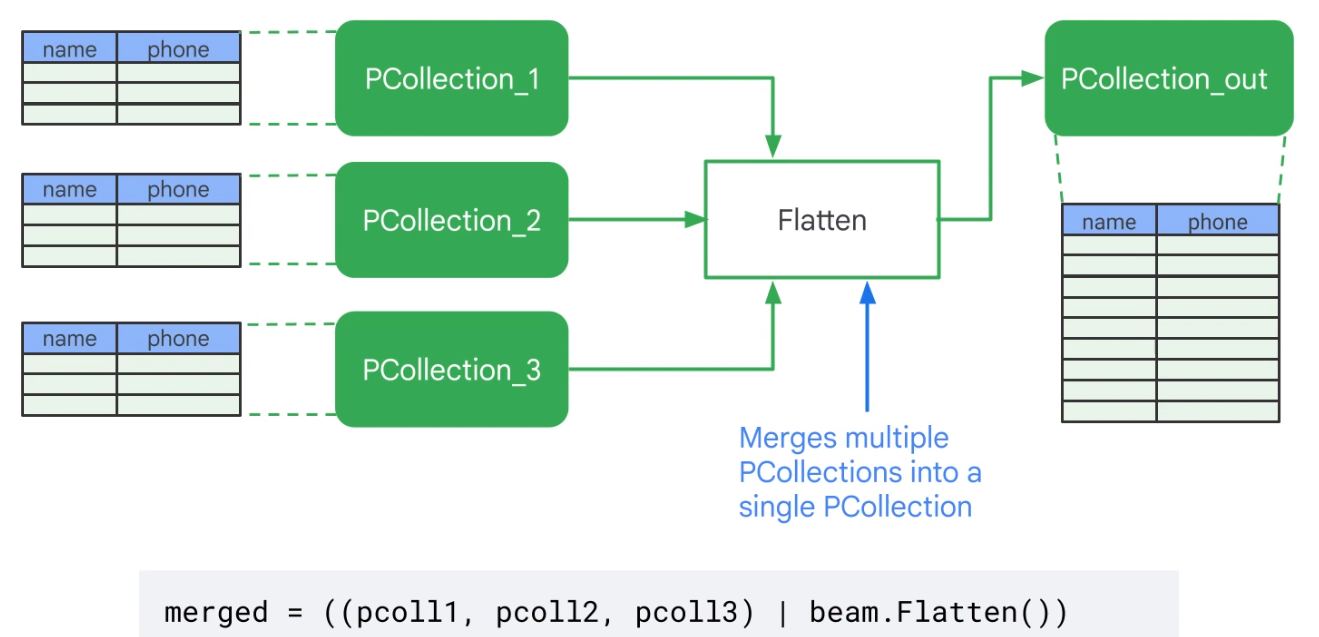
Flatten Merges identical PCollections
Partition Splits PCollections
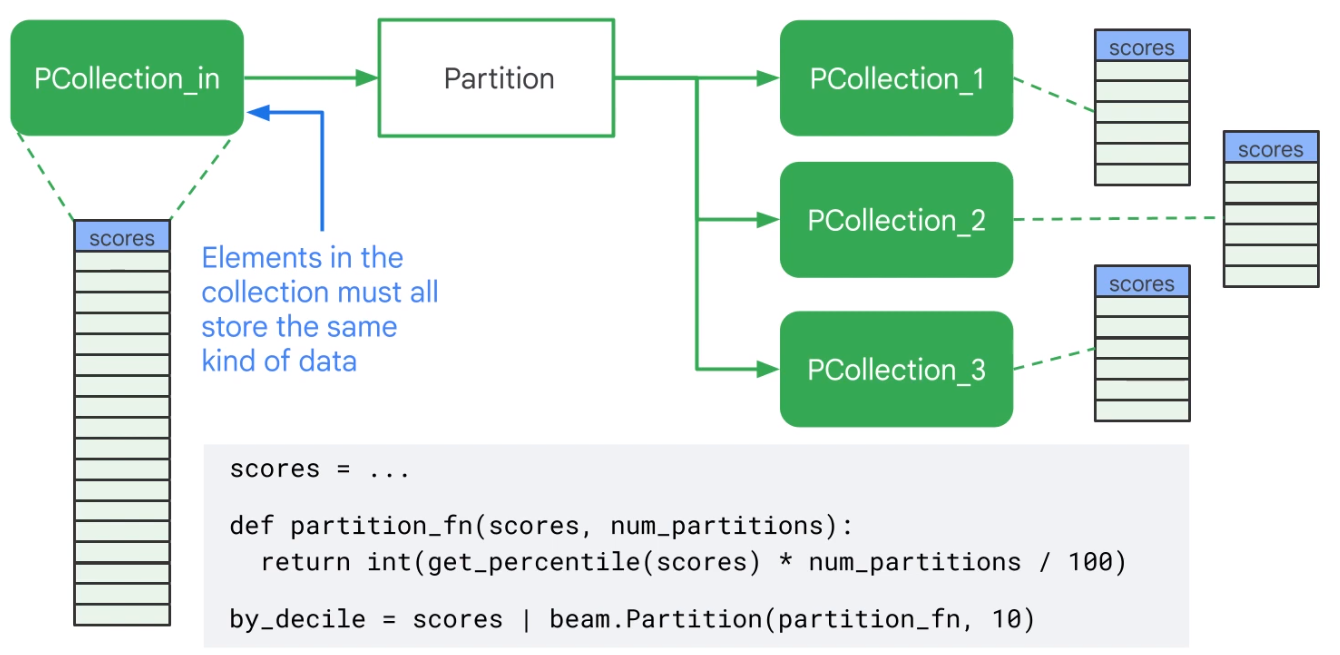
Side-Inputs and Windows
Side-Input
A side-input is an input the do-function can access every time it processes an element of the inputP collection.
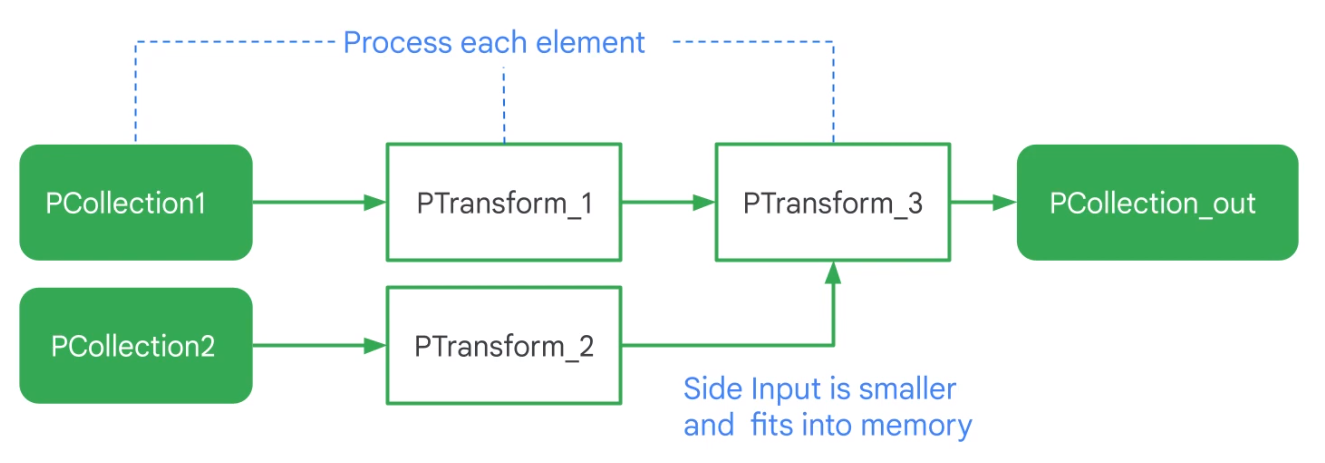
words = ...
def filter_using_length(word, lower_bound, upper_bound=floar('inf')):
if lower_bound <= len(word) <= upper_bound:
yield word
small_words = words | 'small' >> beam.FlatMap(filter_using_length, 0, 3)
# Side input
avg_word_len = (words
| beam.Map(len)
| beam.CombineGlobally(beam.containers.MeanCombineFn())
larger_than_average = (words | 'large' >> beam.FlatMap(
filter_using_length,
lower_bound=value.AsSingleton(avg_word_len)))
Window
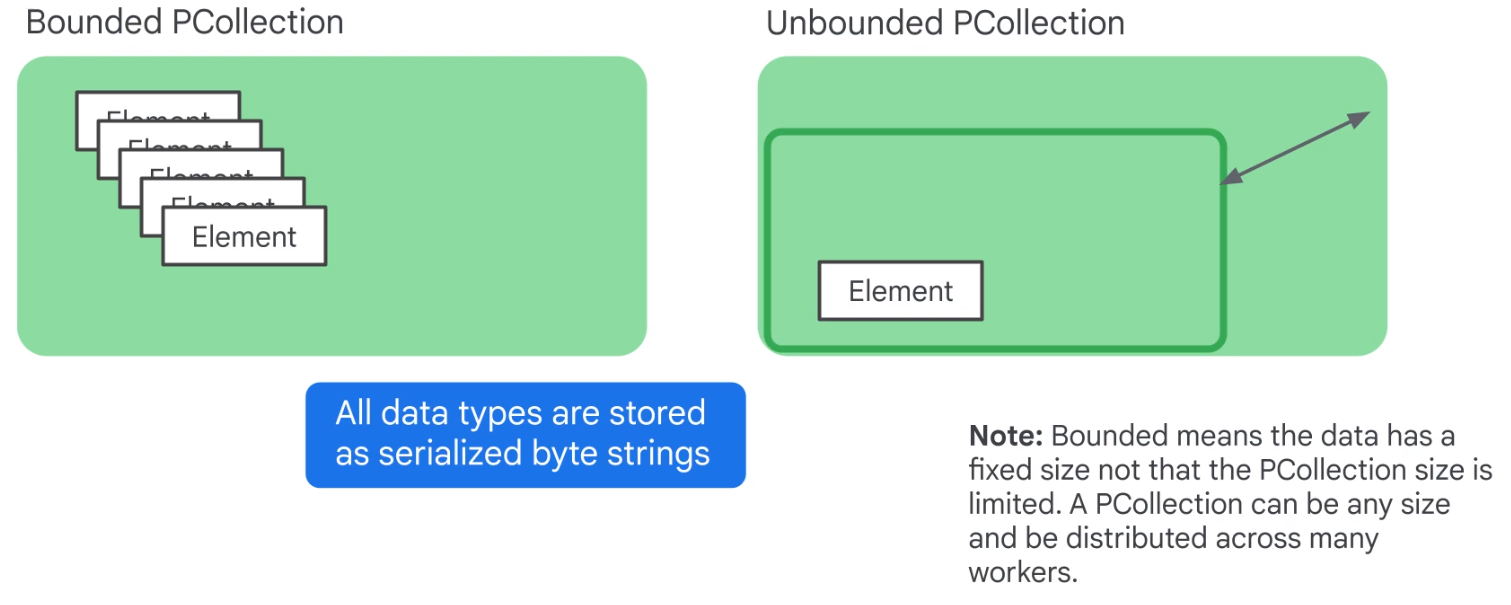
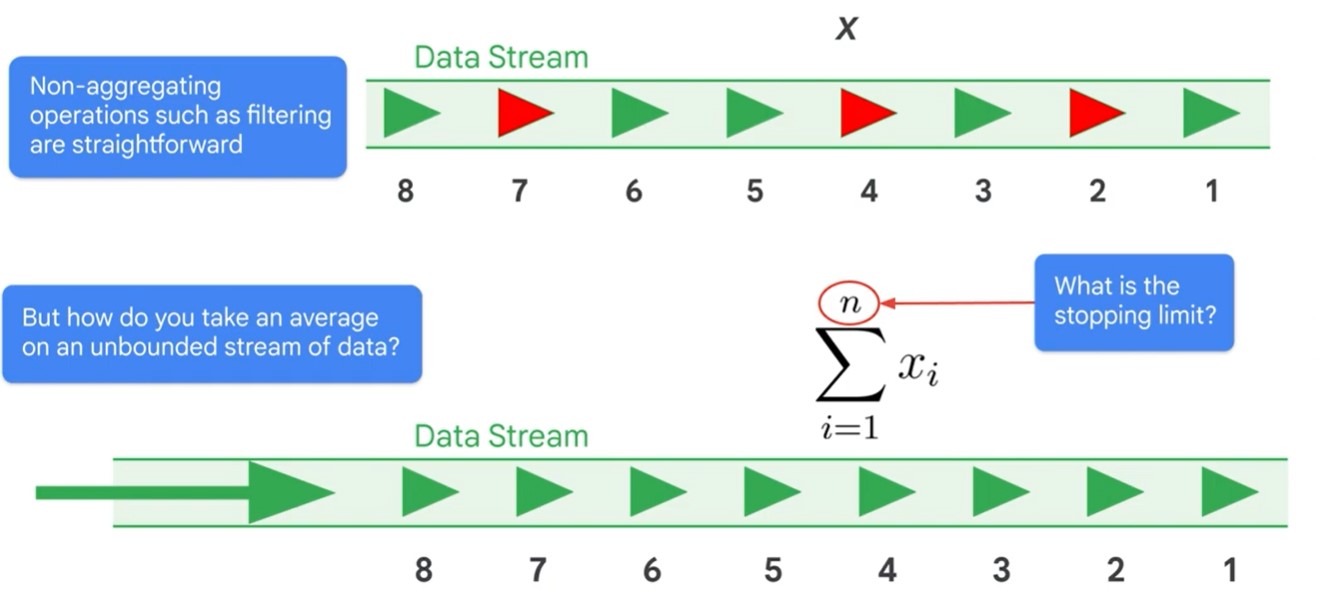
Unbounded PCCollection not useful for Streaming data.
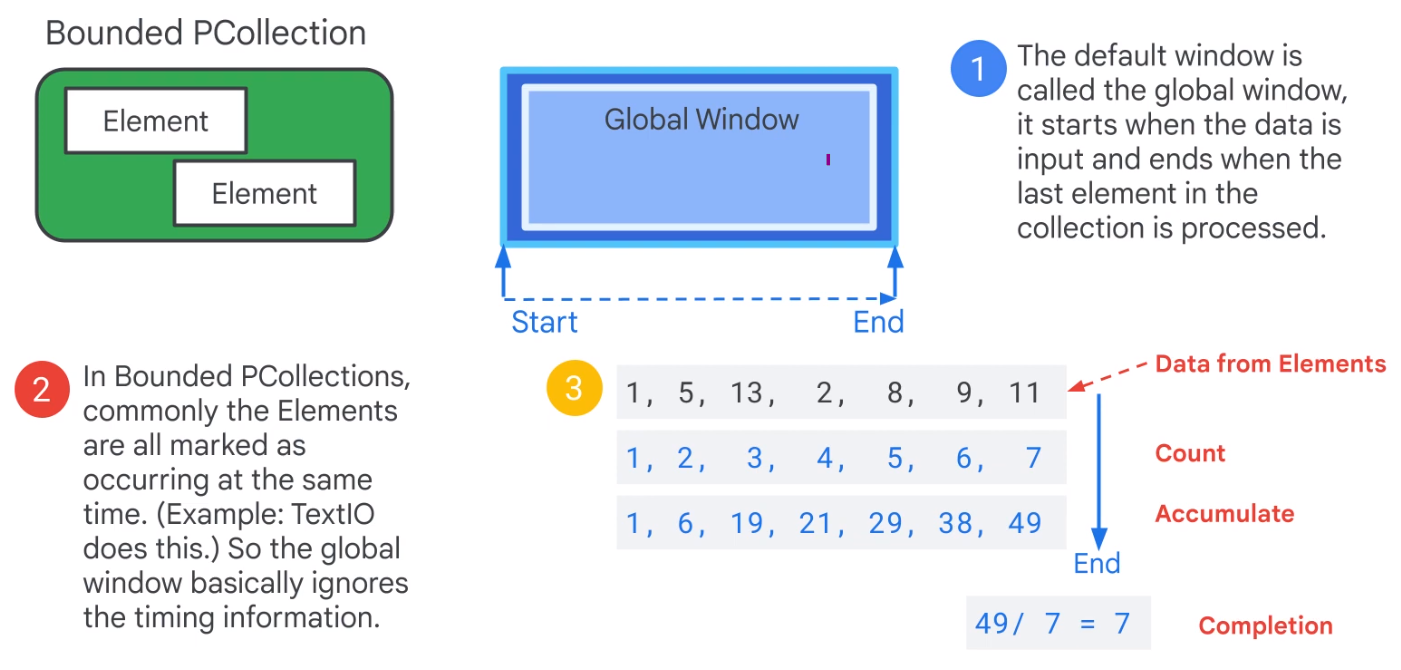
Use time windows.
lines = p | 'Create' >> beam.io.ReadFromText('access.log')
windowd_counts = (
lines
| 'Timestamp' >> beam.Map(beam.window.TimestampedValue(X, extract_timestamp(x)))
| 'Window' >> beam.WindowInto(beam.window.SlidingWindows(60,30))
| 'Count' >> (beam.CombineGlobally(beam.combiners.CountCombineFn()).without_defaults())
)
windowed_counts = windowed_counts | beam.ParDo(PrintWindowFn())
Templates
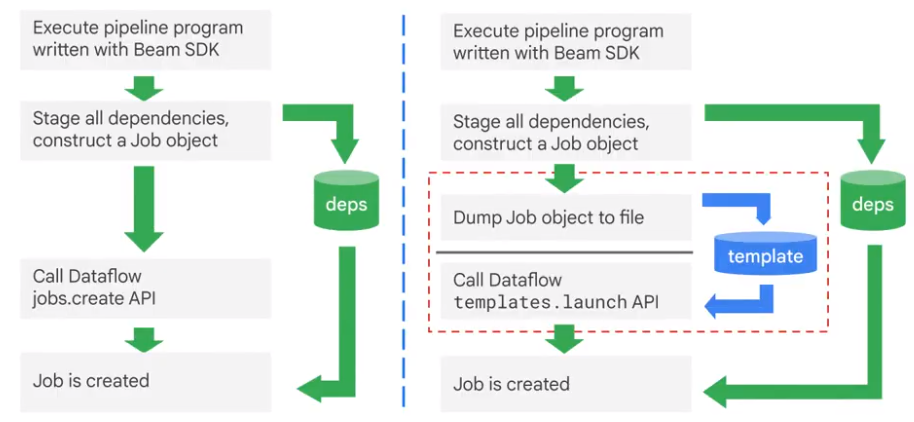
Separate developer from user.
Create own Template

Each template has metadata:

Dataflow SQL
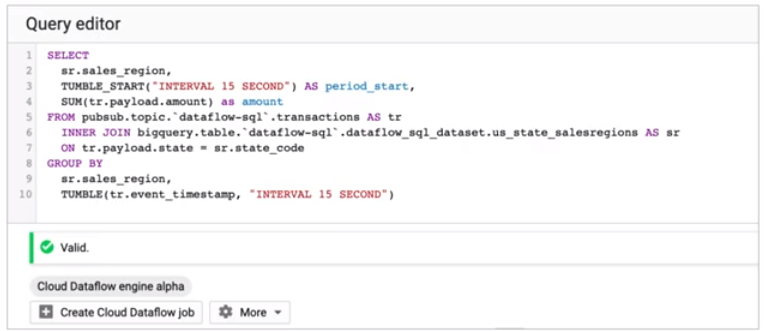
Streaming Data Challenges
- Scalability
- Fault Tolerance
- Model (Sreaming, Repeated Batch)
- Timing
- Aggregation
Message Ordering
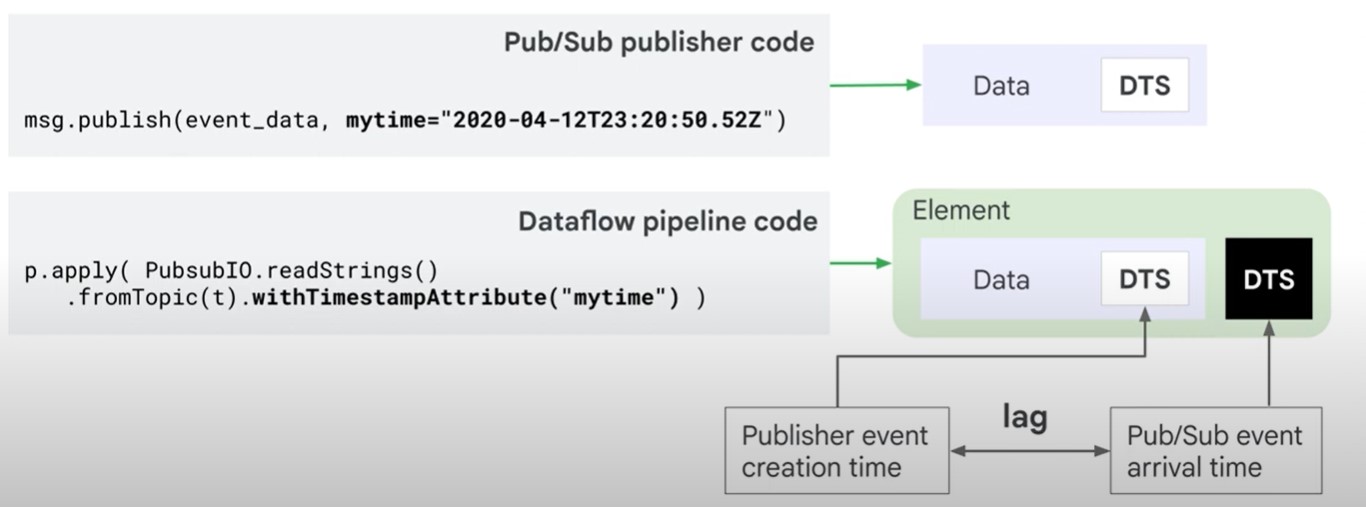
Timestamps can be modified
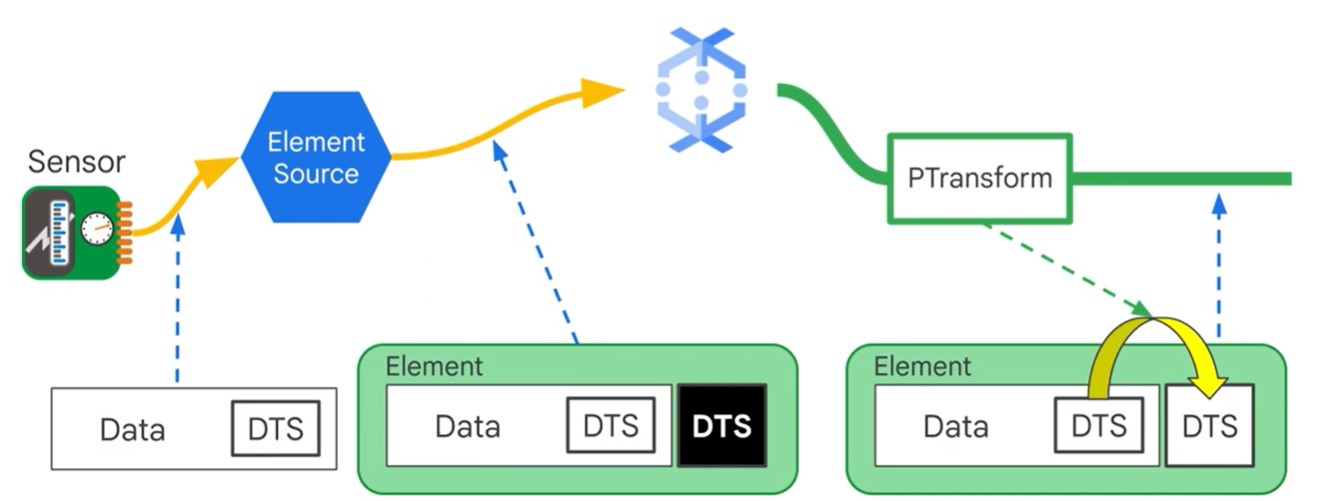
unix_timestamp = extract_timestamp_from_log_entry(element)
yield beam.window.TimestampedValue(element, unix_timestamp)
Duplication
msg.publish(event_data, ,myid="34xwy57223cdg")
p.apply(PubsubIO.readStrings().fromTopic(t).idLabel("myid"))
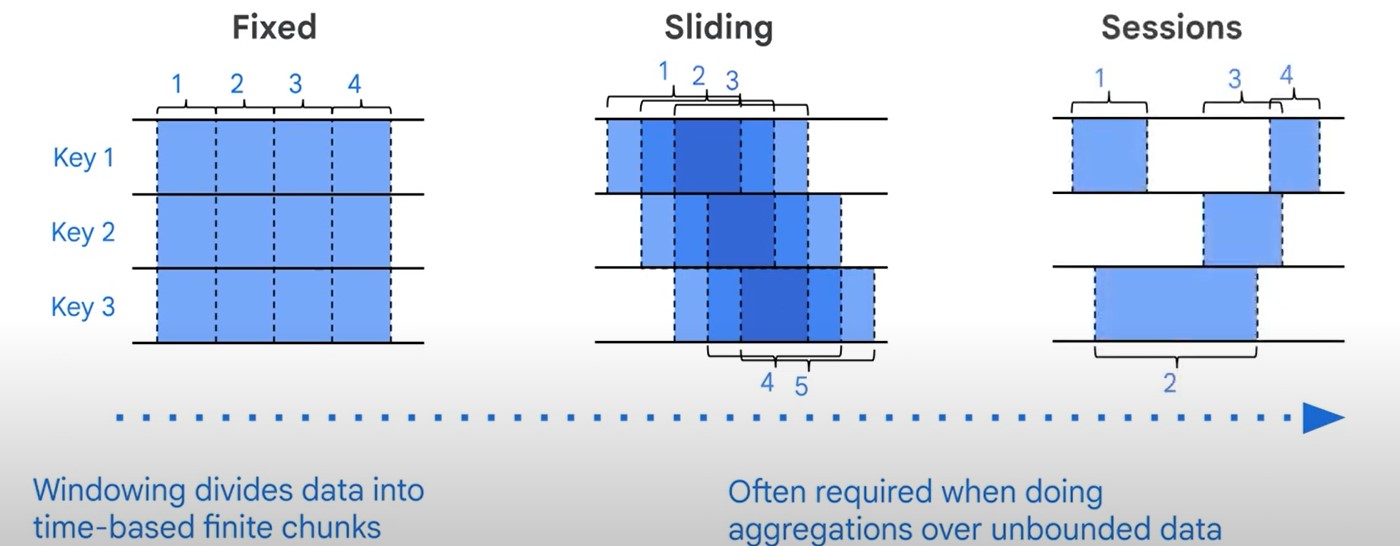
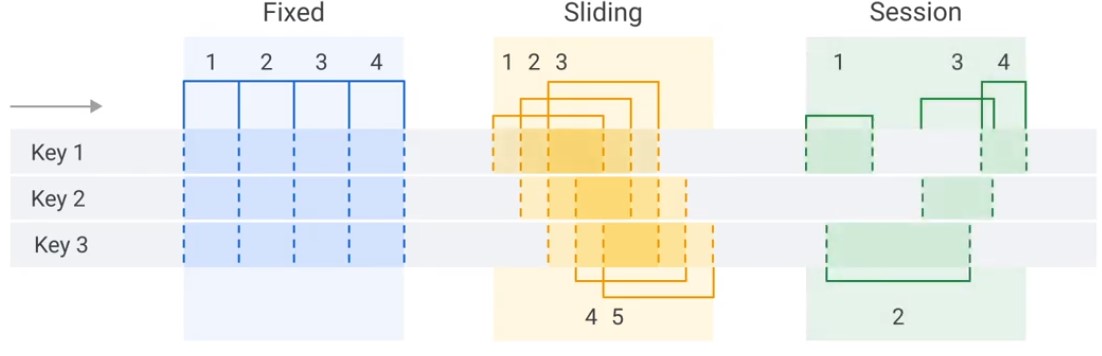
Windowing
- Fixed
- Sliding
- Session
Fixed
from apache_beam import window
fixed_window_items = (items | 'window' >> beam.WindowInto(window.FixedWindows(60)))
Sliding
from apache_beam import window
fixed_window_items = (items | 'window' >> beam.WindowInto(window.SlidingWindows(30,5)))
Session
from apache_beam import window
fixed_window_items = (items | 'window' >> beam.WindowInto(window.Sessions(10*60)))
Pipeline Processing
No Latency
Latencies (Watermark)
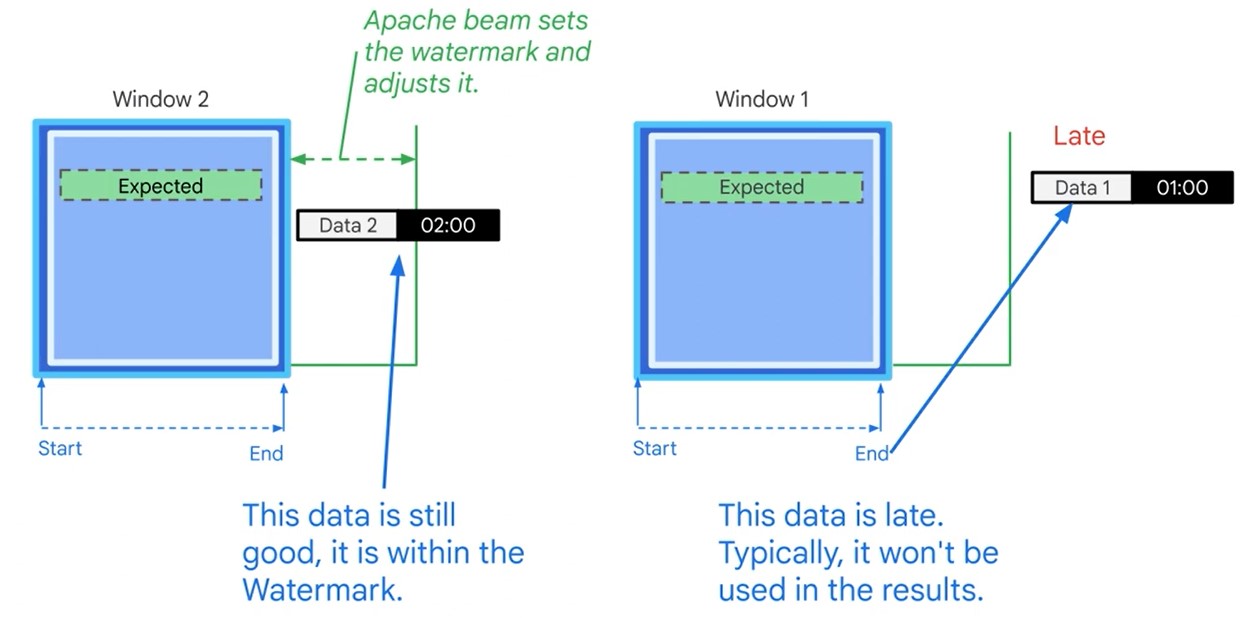
Late messages won’t be processed. You can decide to re-read the dataset.
Triggers
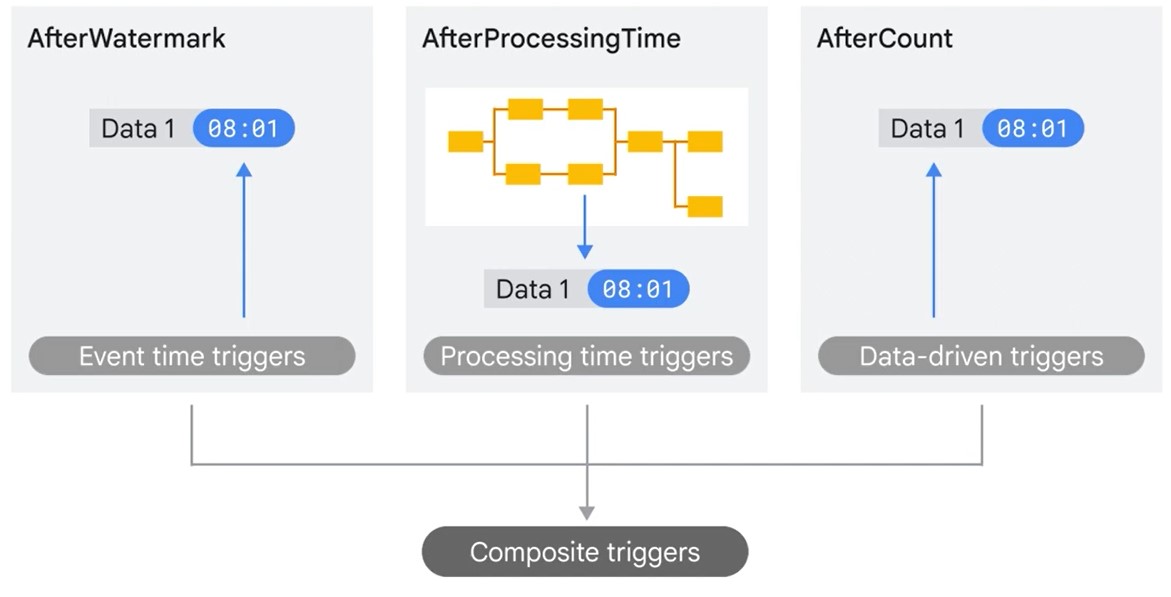
Allow late Data past the Watermark
pc = [Initial PCollection]
pc | beam.WindowInto(
FixedWindow(60),
trigger=trigger_fn,
accumulation_mode=accumulation_mode,
timestamp_combiner=timestamp_combiner,
allowed_lateness=Duration(seconds=2*24*60*60)) # 2 days
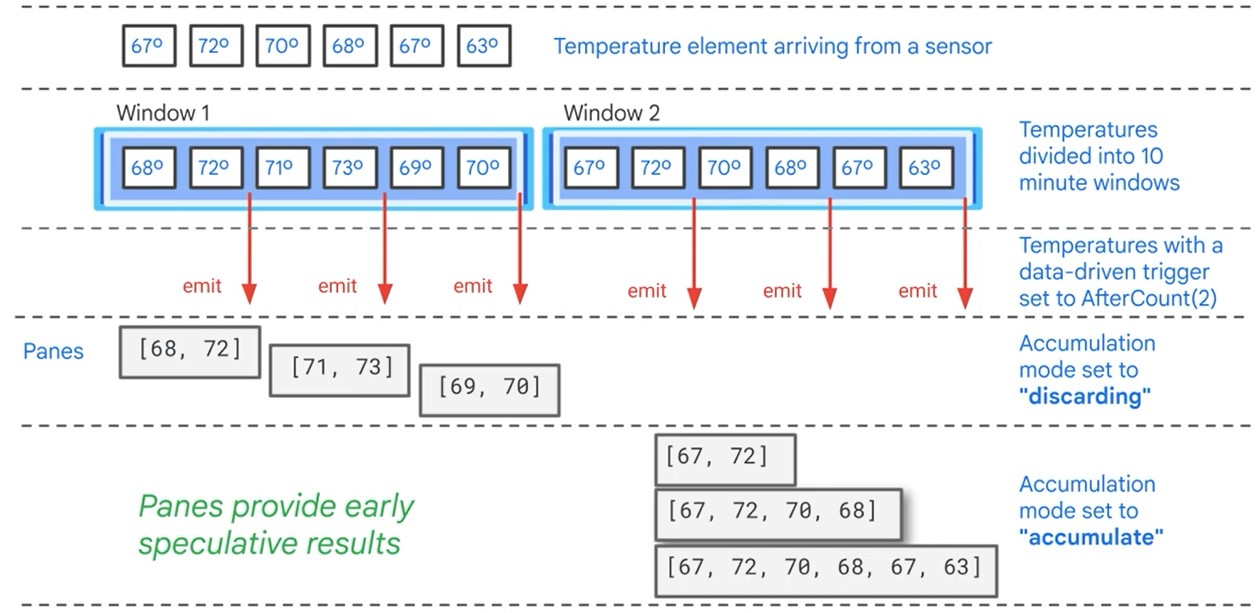
Accumulation Mode
Apache Beam
- Programming model for bath and streaming data pipelines.
- Pipelines can run locally or other backend servers
- A Runner is used to execute you pipeline
Beam Portability
Provide portability framework for data pipelines.
- Portability Framework
- Language agnostic
- Interoperability layer = Portability API
- Every runner works with every language
- Configurable worker environment
- Multi-language pipelines
- Cross-language transforms
- Faster delivery of new features
Dataflow Runner V2
Container Environment
Custom Container
- Apache Beam SDK 2.25.0 or later
- Docker is required, to test pipeline locally
Create Dockerfile
$ from apache/beam_python3.8_sdk:2.25.0
$ env my_file_name=my_file.txt
$ copy path/to/myfile/$MY_FILE_NAME ./
Build Image
$ export PROJECT=my-project-id
$ export REPO=my-repository
$ export TAG=my-image-tag
$ export REGISTRY_HOST=gcr.io
$ export IMAGE_URI=$REGISTRY_HOST/$PROJECT/$REPO:$TAG
$ gcloud builds submit --tag $IMAGE_URI
$ docker build -f Dockerfile -t $IMAGE_URI ./
$ docker push $IMAGE_URI
Launch Job
$ python my-pipeline.py \
--input=INPUT_FILE \
--output=OUTPUT_FILE \
--project=PROJECT_ID \
--region=REGION \
--temp_location=TEMP_LOCATION \
--runner=DataflowRunner \
--worker_harness_container_image=$IMAGE_URI
Cross-Language Transforms
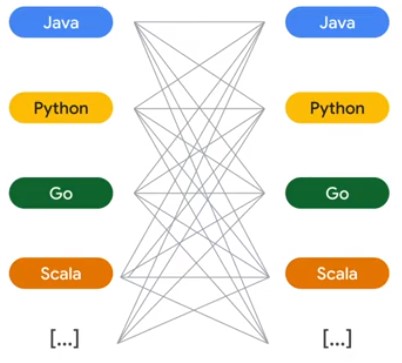
from apache_beam.io.kafka import ReadFromKafka
with beam.Pipeline(options=<You Beam PipelineOptions object>) as p:
p
| ReadFromKafka(
consumer_config={'bootstrap.servers':'Kafka bootstrap servers list>'},
topics=[<List of Kafka topics>])
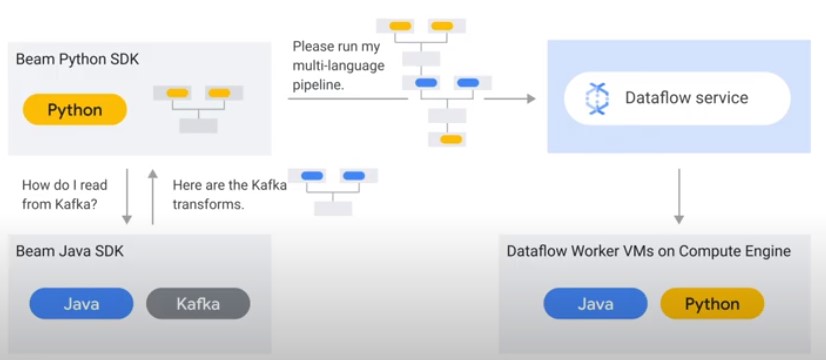
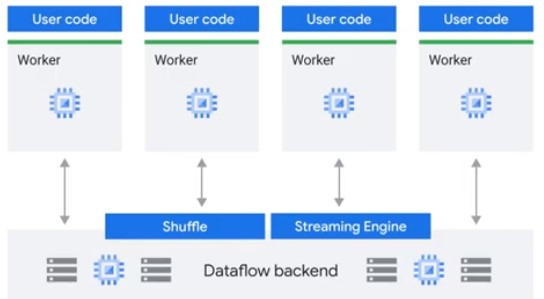
Separate Compute and Storage
Dataflow allows executing Apache Beam Pipelines on Google Cloud.
Dataflow Shuffle Service
Only for Batch pipelines. Faster execution time.
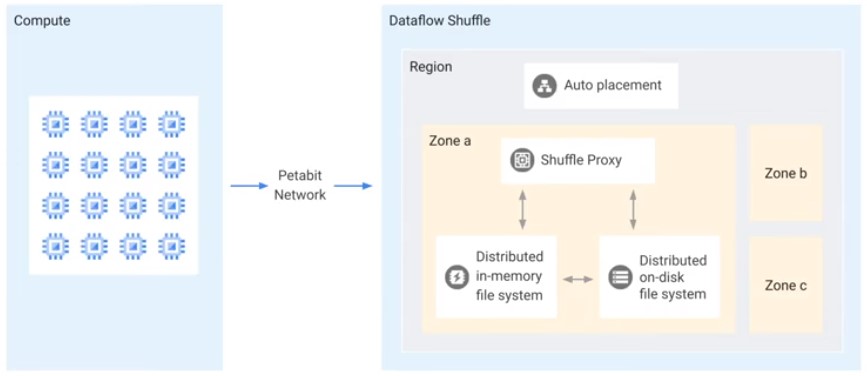
Dataflow Streaming Engine
For Streaming Data pipelines. Less CPU and Memory.
Flexible Resource Scheduling (FlexRS)
Reduce cost of batch processing pipelines
Execution within 6 hours. For non-time critical workflows.
IAM
Job Submission
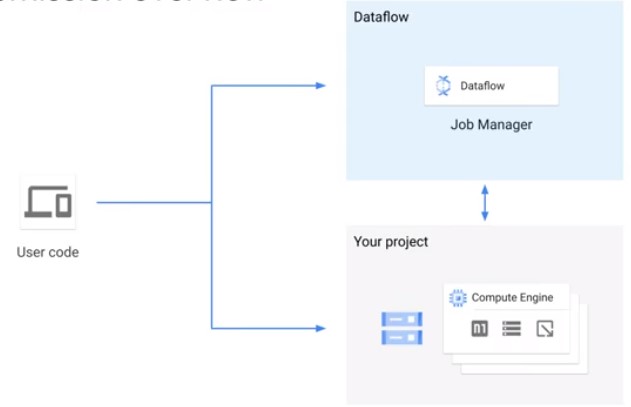
Three Credentials
User roles
- Dataflow Viewer: Read-Only access to DF ressources
- Dataflow Developer: View, Update and Cancel DF jobs
- Dataflow Admin: Create and manage Dataflow jobs
Dataflow Service Account
- Interacts between project and Dataflow
- Used for worker creation and monitoring
- service-
@dataflow-service-producer-prod.iam.gserviceaccount.com - Dataflow Agent role
Dataflow Controller Service Account
- Used by the workers to access resources needed by the pipeline
-
-compute@developer.gserviceaccount.com
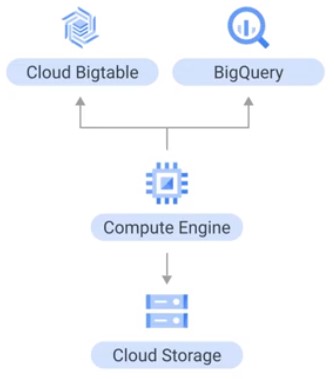
Quotas
CPU
Total number of CPUs consumed in a region.
gcp > IAM > Quota.
IP
Total number of VMs with external IP address in a region.
Persistent Disks
Either HDD, or SSD. set –worker_disk_type flag (pd-ssd).
Batch
- VM to PD ration is 1:1 for batch
- Size if Shuffle on VM: 250 GB
- Size if Shuffle Service: 25 GB
- Override default: –disk_size_gb
Streaming
- Fixed numer of PD
- Default size if shuffle on VM: 400 GB
- Shuffle service: 30 GB
- Amount of disk allocated == Max number of workers
- –max_num_workers
Security
Data Locality
Ensure all data and metadata stays in one region.
- Backend that deploys and controls Dataflow workers.
- Dataflow Service accound talks with regional endpoint.
- Stores and handles metadata about you DF job.
Reasons for regional endpoint.
- Security and complience
- No zone preference: –region $REGION
- Specific region endpoint: –region $REGION – worker_zone $WORKER_ZONE
Shared VPC (Virtual Private Cloud)
- Jobs can run in either VPC of Shared VPC
- Works for both default and custom networks
- Number of VMs is constrained by subnet IP block size
- Dataflow service account needs Compute Network User role in host project
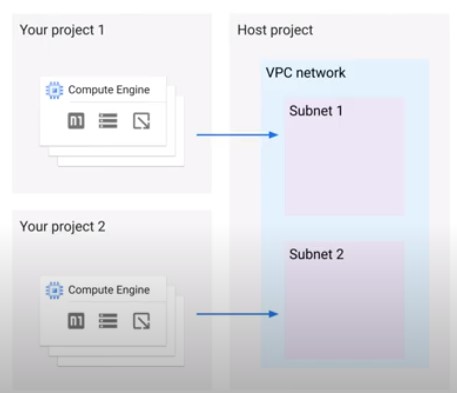
- –network default
Private IPs
- Secure you data processing infrastructure
- Pipeline cannot access the internet and other Google Cloud networks
- Network must have Pricate Google Access to reach Google Cloud APIs and services
- –subnetwork regions/$REGION/subnetworks/$SUBNETWORK
–no_use_public_ips
CMEK
Customer Managed Encryption Key. By default, Google manages key encryption. Customers can use symmetric CMEK keys stored in Google Cloud managed key service. Job metadata is encrypted with Google encryption.
- –temp_location gs://$BUCKET/tmpt/
–dataflow_kms_key=projects/$PROJECT/locations/$REGION/keyRings/$KEY_RING/cryptoKeys/$KEY </a>
Dataflow Pipelines
Start Dataflow Pipeline
Dataflow Pipeline Coding Examples
Beam Basics (Beam = Batch + Stream)
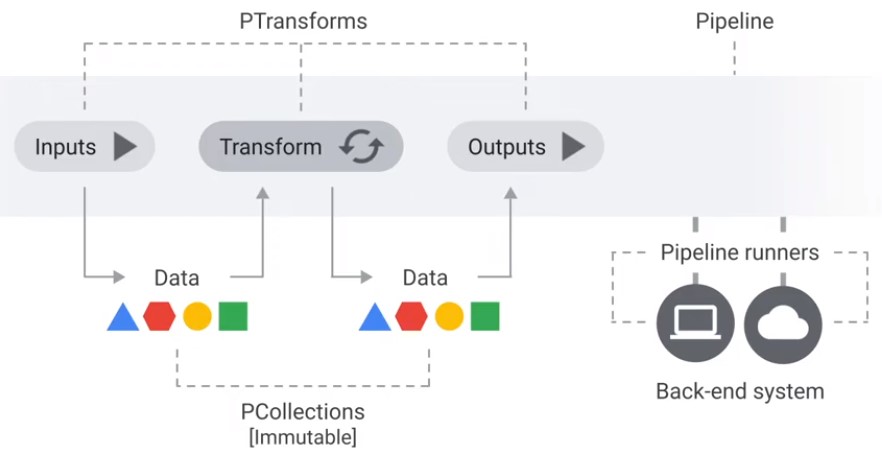
Transforms
| Transformation | Description |
|---|---|
| ParDo | The most-general mechanism for applying a user-defined DoFn to every element in the input collection. |
| Combine | Transforms to combine elements. |
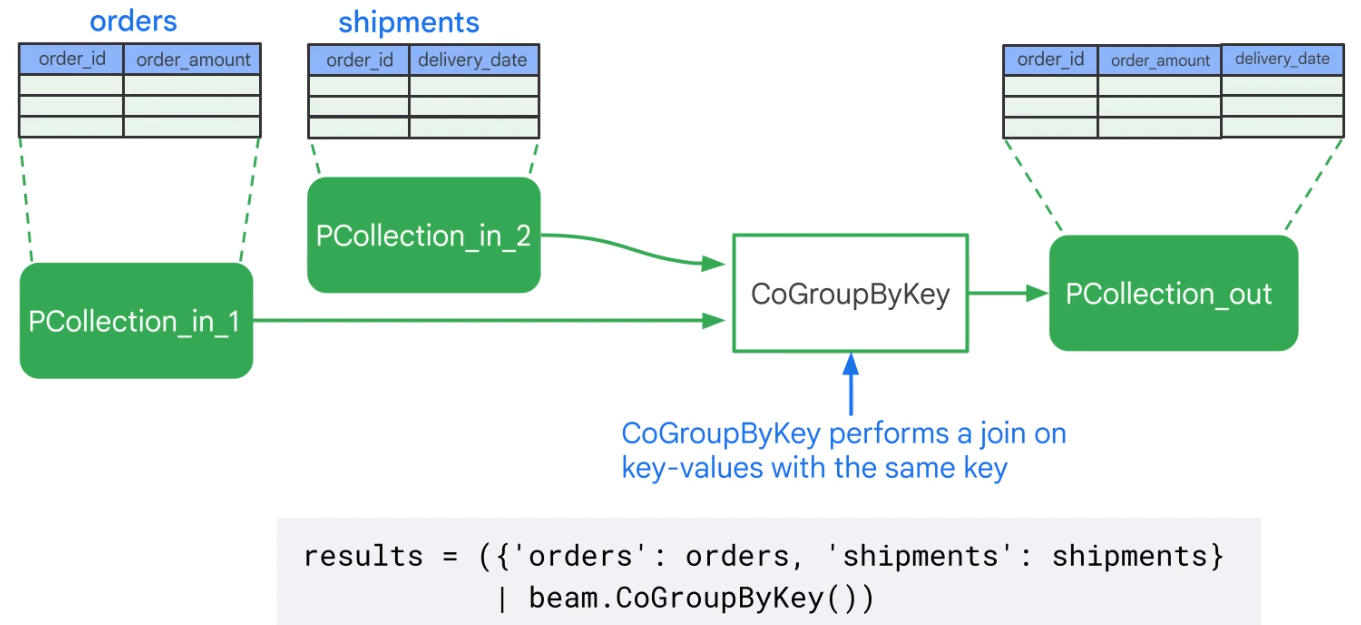
| GroupByKey | Takes a keyed collection of elements and produces a collection where each element consists of a key and all values associated with that key. |
| CoGroupByKey | Takes several keyed collections of elements and produces a collection where each element consists of a key and all values associated with that key. |
| Flatten | Given multiple input collections, produces a single output collection containing all elements from all of the input collections. |
| Partition | Routes each input element to a specific output collection based on some partition function. |
Python Transformation Catalogue
DoFn
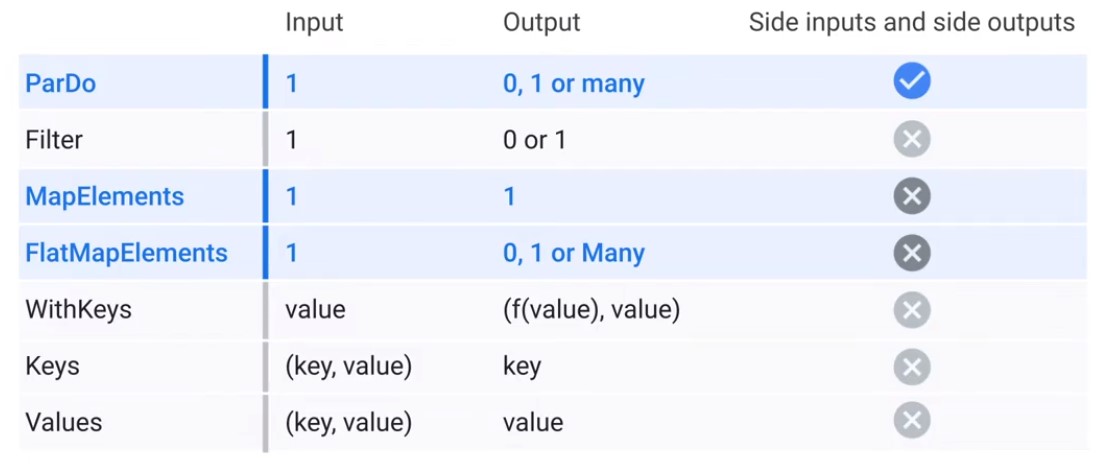
Data Bundles
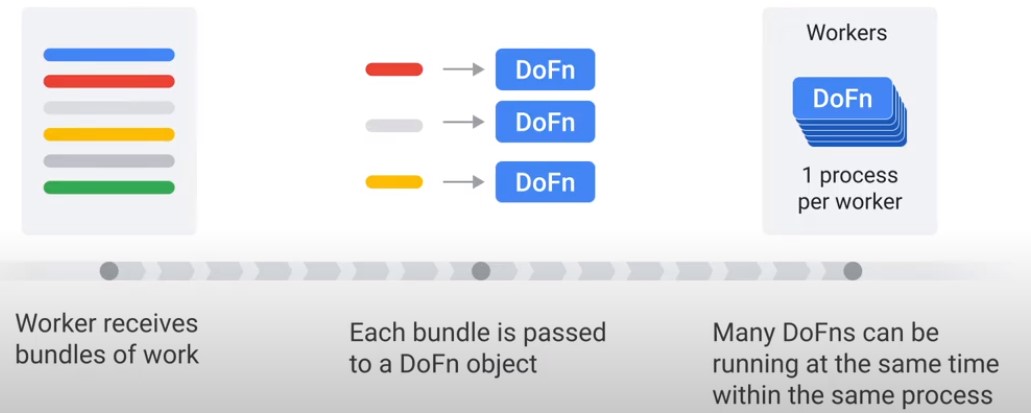
Methods of DoDn
class MyDoFn(bean.DoFn):
def setup(self):
pass
def start_bundle(self):
pass
def process(self,element):
pass
def finish_bundle(self):
pass
def teardown(self):
pass
Lifecycle
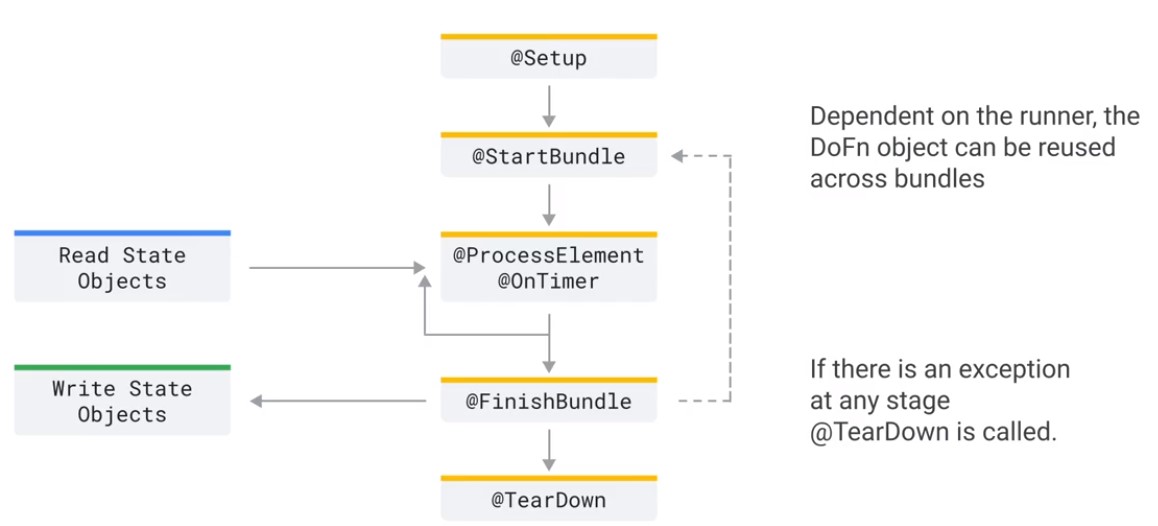
Dataflow Streaming Data
Batch vs Streaming
Out order Stream.

Windows
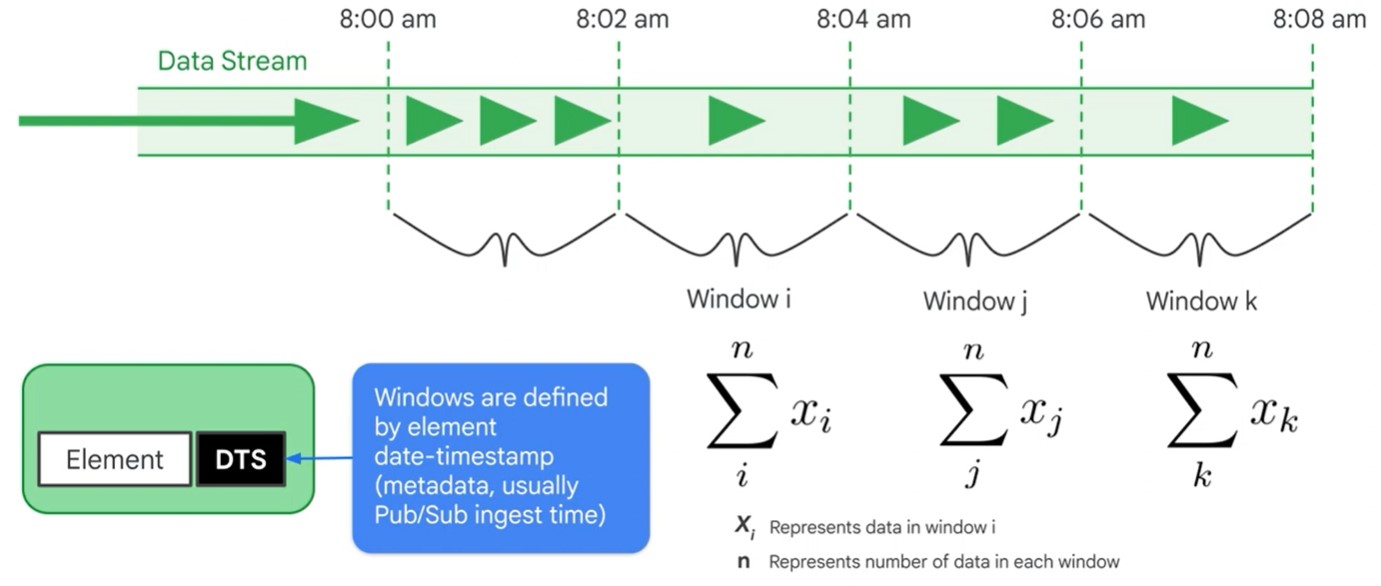
- Windowing divides data into time-based finite chunks.
- Required when doing aggregations over unbound data using Beam primitives (GroupByKey, Combiners).
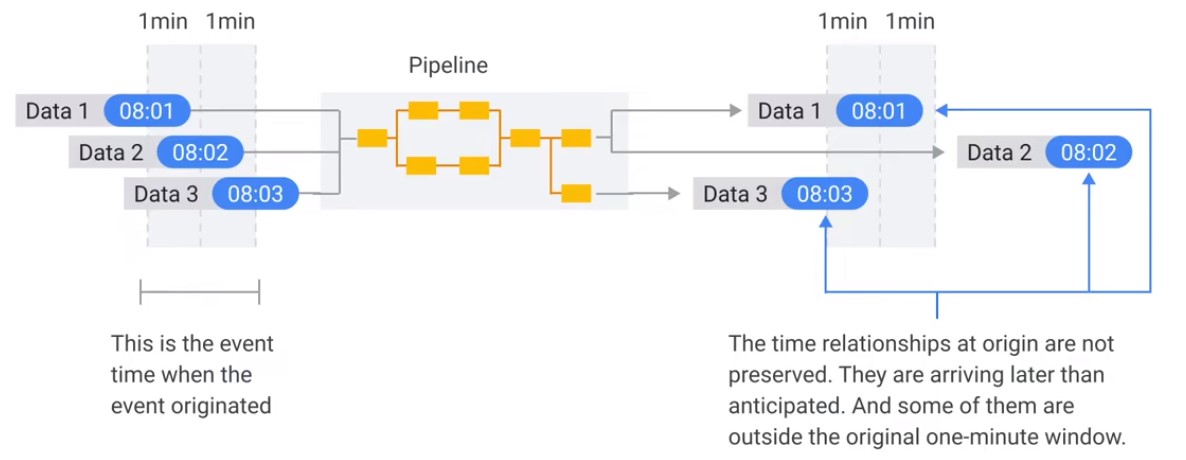
Two Dimensions of Time
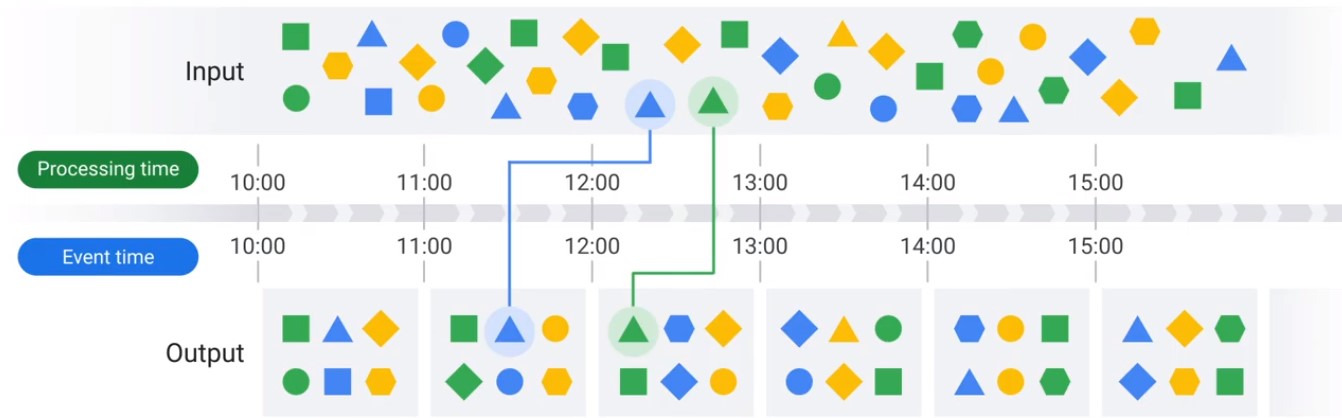
Types of Windows
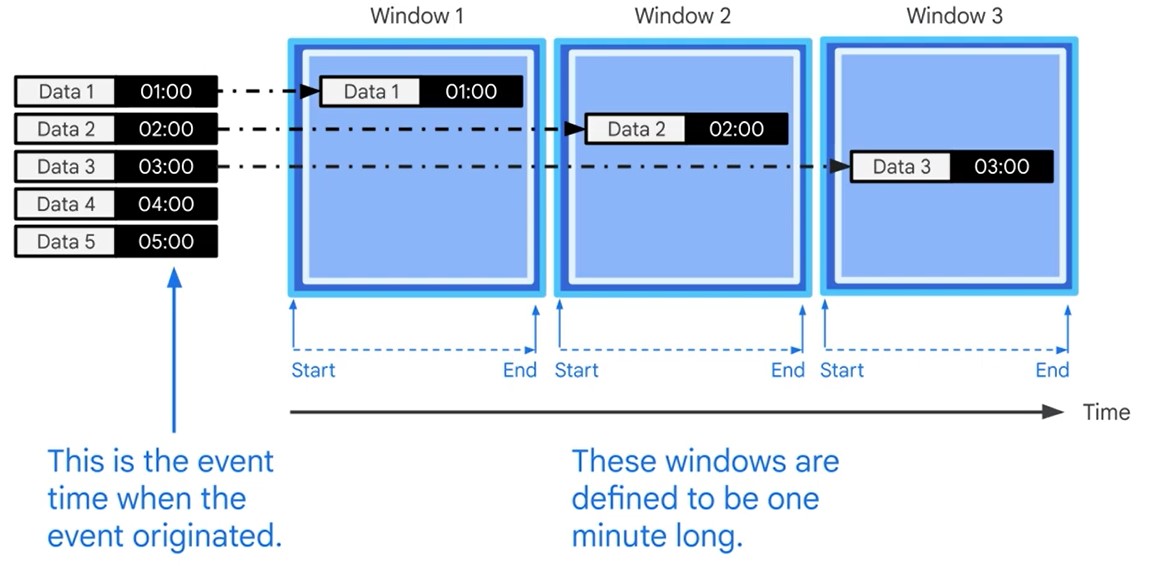
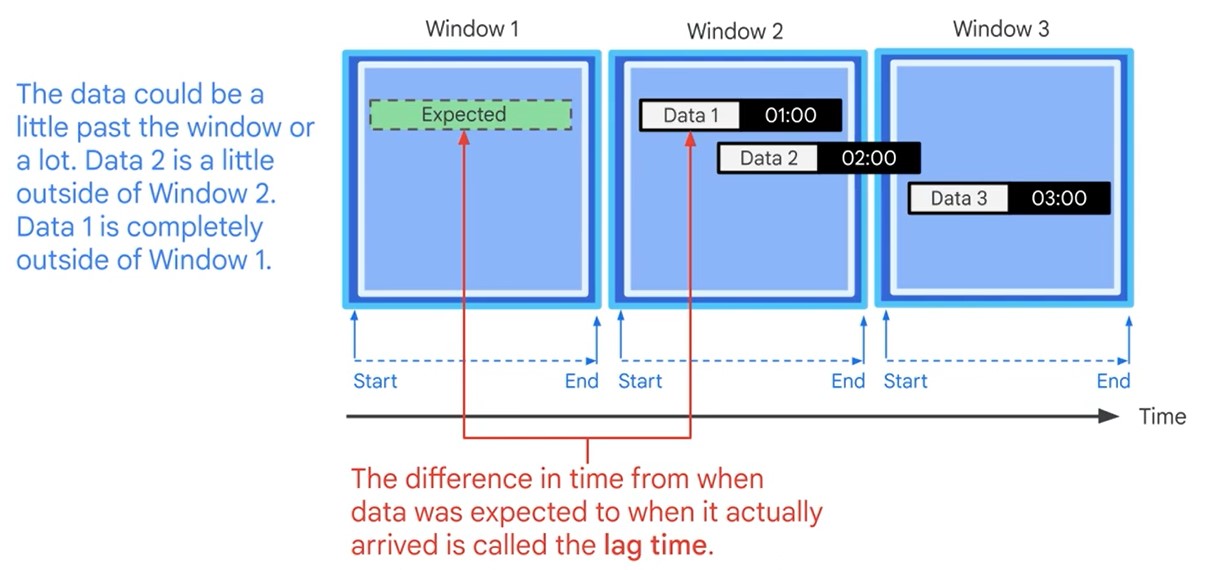
Watermarks
Latency Problem. When to close the Window?
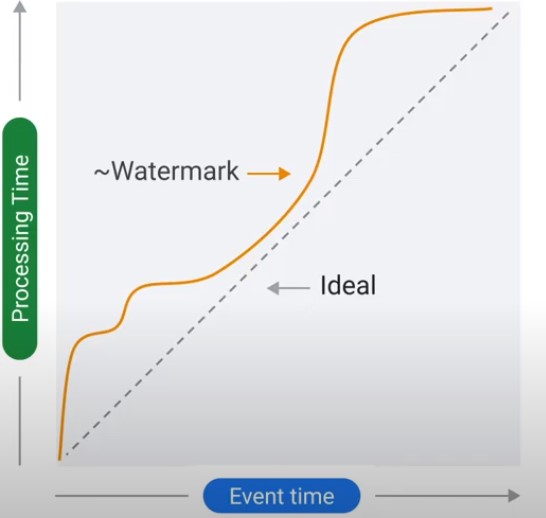
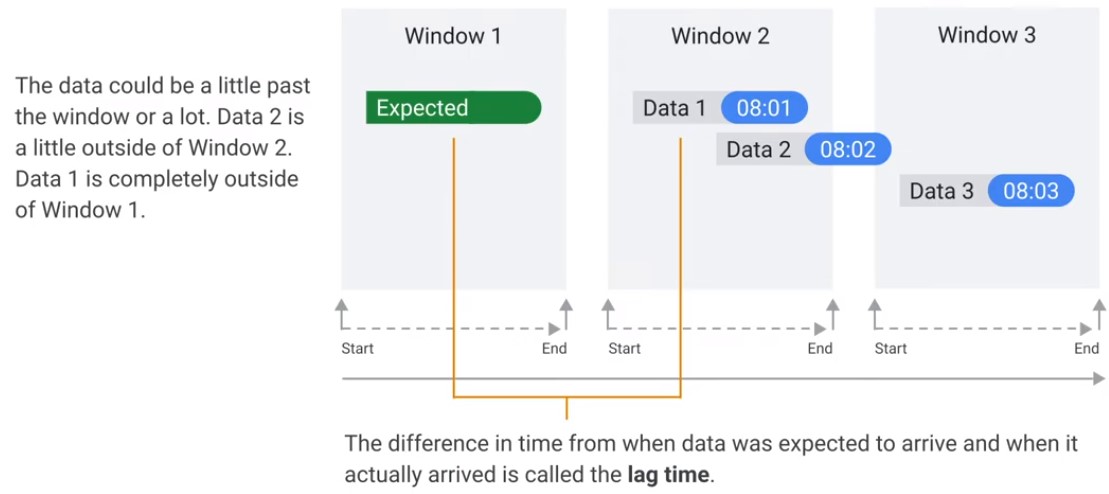
Time Lag
Data is only late when it exceeds the watermark.
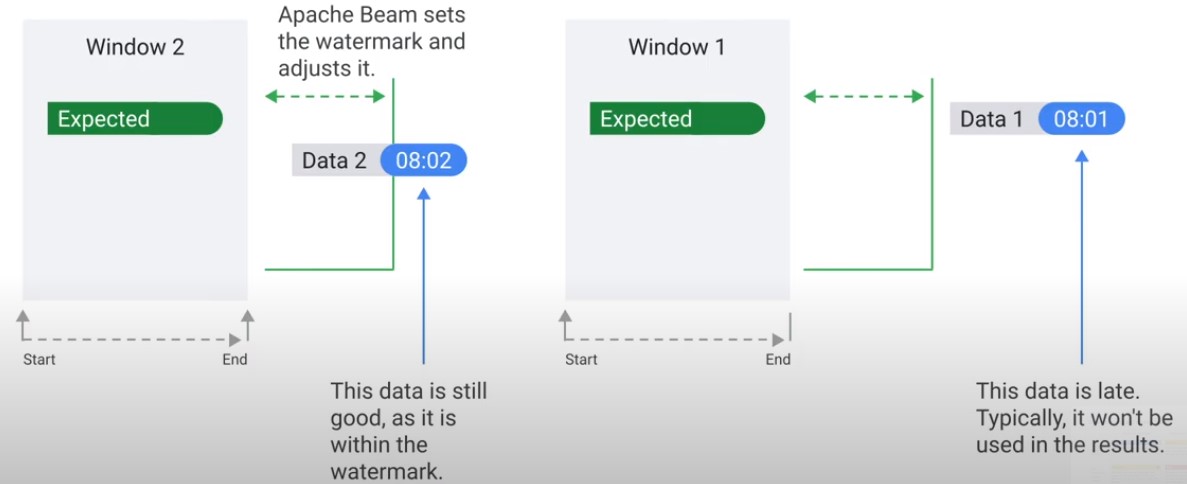
Observe Watermark
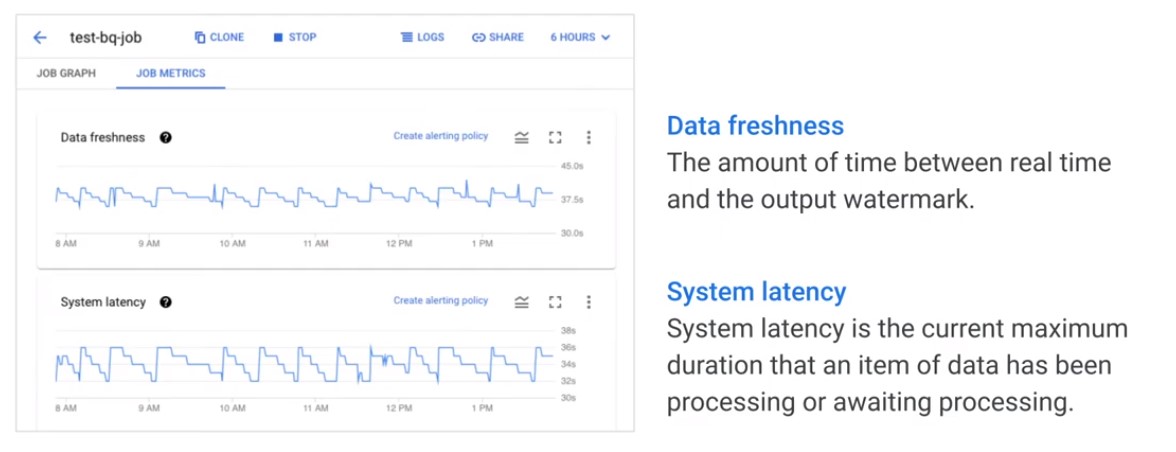
- Data Freshness: The amount of time between real time and the output watermark.
- System Latency: System latency is the current maximum duration that an item of data has been processing.

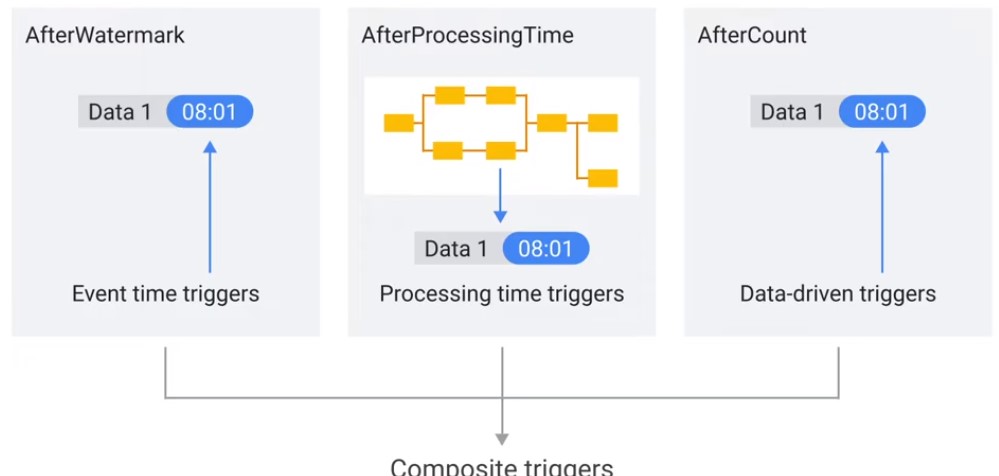
Triggers
Decide when to close window, even if late data has not arrived.
- Event Time (After Watermark)
- Processing Time (After Processing Time)
- Composite
- Data-driven (After Count)
Custom Triggers
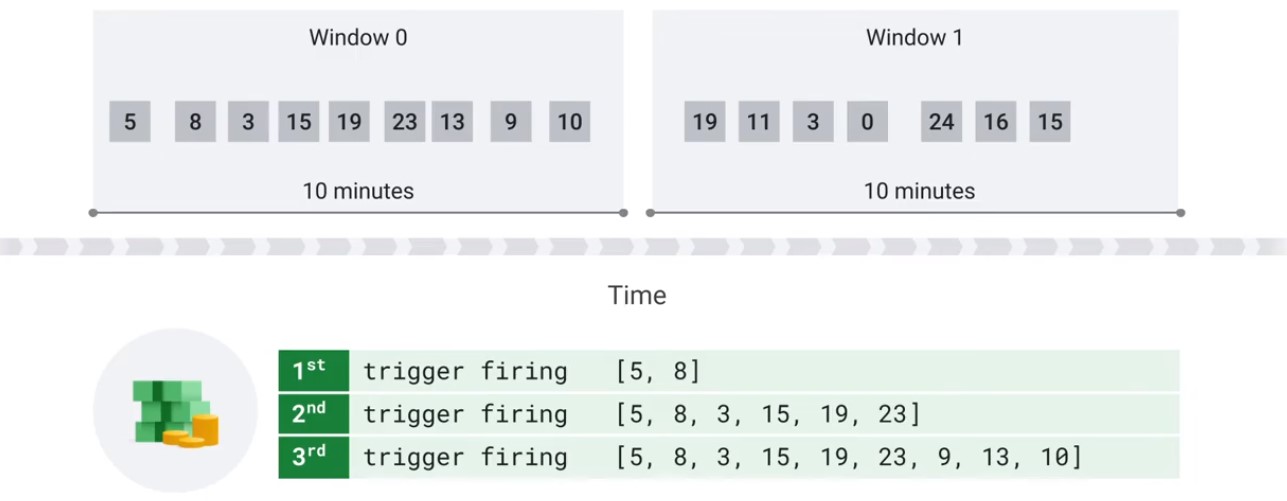
Accumulate Mode
Accumulate
pcollection | Windowinto(
SlidingWindows(60,5),
trigger=AfterWatermark(
early=AfterProcessingTime(delay=30),
late=AfterCount(1))
accumulation_mode=AccumulationMode.ACCUMULATING)
Discard
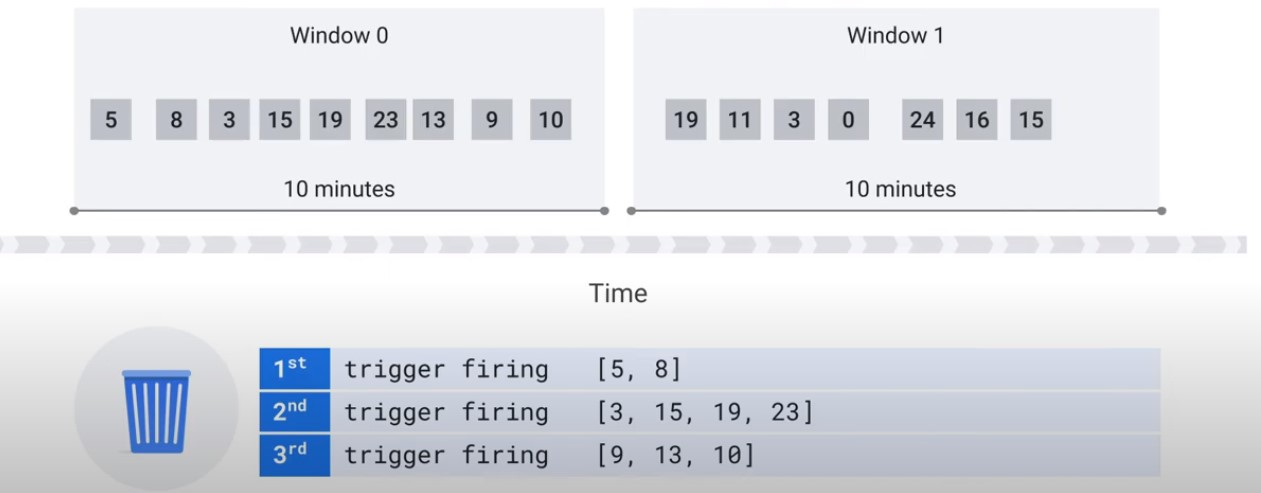
pcollection | WindowInto(
FixedWindow(60),
trigger=Repeatedly(
AfterAny(
AfterCound(100),
AfterProcessingTime(1*60))),
accumulation_mode=AccumulationMode.DISCARDING,
allowed_lateness=Duration(seconds=2*24*60*60))
Sources and Sinks
- Source: Read Data
- Bounded: Batch Data (Read Data in Bundles)
- Unbounded: Stream Data
- Sink: Write Data (PTransform Write)
Beam Data Sinks
Java
@AutoValue
public abstract static class Write<T> extends
PTransform<PCollectoin<T>,WriteResult> {
Python
class WriteToPubSub(PTransform):
Text IO
Reading
Java
Pipeline
.apply(
"Read from source",
TextIO
.read()
.from(options
.getInputFilePattern()))
Reading with Filenames
p.apply(
FileIO
.match()
.filepattern("hdfs://path/to/*.gz"))
.apply(
FileIO
.readMatches().withCompression(Compression.GZIP))
.apply(
ParDo.of(
mew DoDn<FileIO.ReadableFile, String>() {
@ProcessElement
public void process(
@Element FileIO.ReadableFile file) {
LOG.info("File Metadata resourceId is {} ",
file.getMetadata().resourceId());
}
}));
Processing Files as they arrive
p.apply(
FileIO.match()
.filepattern("...")
.continuously (
Duration.standardSeconds(30),
Watch.Growth.afterTimeSinceNewOutput(
Duration.standardHours(1))));
Contextual Text reading
PCollection<Row> records1 =
p.apply(ContextualTextIO.read().from("..."));
PCollection<Row> records2 =
p.apply(ContextualTextIO.read()
.from("/local/path/to/files/*.csv")
.withHasMultilineCSVRecors(true));
PCollection<Row> records3 =
p.apply(ContexturalTextIO.read()
.from("/local/path/to/files/*")
.watchForNewFiles(
Duration.standardMinutes(1),
afterTimeSinceNewOutput(
Duration.standardHours(1))));
Python
pcoll1 = (pipeline
| 'Create' >> Create([file_name])
| 'ReadAll' >> ReadAllFromText())
pcoll2 = pipeline | 'Read' >> ReadFromText(file_name)
Reading with Filenames
with beam.Pipeline() as p:
readable_files = (
p
| fileio.MatchFiles ('hdfs://path/to/*.txt')
| fileio.ReadMatches()
| beam.Reshuffle())
files_and_contens = (
readable_files
| beam.Map(lambda x: (x.metadata.path,
x.read_utf8)))
Processing Files as they Arrive
with beam.Pipeline() as p:
readable_files = (
p
| beam.io.ReadFromPubSub(...)
... #<Parse PubSub Message and Yield Filename>
)
files_and_contents = (
readable_files
| ReadAllFromText())
Writing
Java
csv.appy(
"Write to storage",
TextIO
.write()
.to(Options
.getTextWritePrefix())
.withSuffix(".csv"));
Text Writing with Dynamic Destinations
PCollection<BankTransaction> transactions = ...;
transactions.apply(FileIO.<TransactionType,
Transaction>writeDynamic()
.by(Transaction::getTypeName)
.via(tx -> tx.getTypeName().toFields(tx),
type -> new CSVSink(type.getFieldNames()))
.to(".../path/to/")
.withNaming(type -> defaultNaming(
type + "-transactions", ".csv"));
Python
transformed_data
| 'write' >> WriteToText(
known_args.output, coder=JsonCoder()))
Text Writing with Dynamic Destinations
(my_pcollection
| beam.io.fileio.WriteToFiles(
path='/my/file/path',
destination=lamba record: 'avro'
if record ['type'] == 'A' else 'csv',
sink = lamda dest: AvroSink()
if fest == 'avro' else CsvSink(),
file_naming = beam.io.fileio
.destination_prefix_naming()))
### BigQuery IO
#### Reading
##### Java
PCollection<Double> maxTemperatures =
p.apply(
BigQueryIO.read(
(SchemaAndRecord elem) -> (Double)
elem.getRecord()
.get("max_temperature"))
.fromQuery(
"select max_temperature from
`clouddataflow-readonly.samples.weather_stations`")
.usingStandardSql()
.withCoder(DoubleCoder.of()));
Reading with BiqQuery Storage API
PCollection<MyData> rows =
pipeline.apply("Read from BitQuery table",
BigQueryIO.readTableRows()
.from(
String.format("%s:%s,%s,
project, dataset, table))
.withMethod(Method.DIRECT_READ)
//.withRowRestriction
.withSelectedFields(
Arrays.asList(..."string_...","Int64...")))
.apply("TableRows to MyData",
MapElements.into(
TypeDescriptor.of(MyData.class))
.via(MyData::fromTableRow))
Python
max_temperatures = (
p
| 'QueryTableStdSQL' >> beam.io.ReadFromBigQuery(
query = 'select max_temperature from '\
`clouddataflow-readonly.samples.weather_stations`',
use_standard_sql=True)
| beam.Map(lambda elem: elem['max_temperature']))
Writing
Java
Dynamic Destinations
pc.apply(BigQueryIO.<Purchase>write(tableSpec)
.useBeamSchema()
.to((ValueInSingleWindow<Purchase> purchase) -> {
return new TableDestition(
"project:dataset-" +
purchase.getValue().getUser() +
":purchases","");
});
Python
def table_fn(element, fictional_characters):
if element in fictional_characters:
return 'my_dataset.fictional_quotes'
else:
return 'my_dataset.real_quotes'
quotes | 'WriteWithDynamicDestination' >>
beam.io.WriteToBigQuery(
table_fn,
schema=table_schema,
table_side_inputs=(fictional_characters_view, ),
...)
Pub/Sub IO
Java
pipline
.apply("Read PubSub Messages",
PubsubIO
.readStrings()
.fromTopic(options.getInputTopic()))
.apply(
Window.into(
FixedWindows.of(
Duration.standardMinutes(
options.getWindowSize()))));
Python
class GroupWindowsIntoBatches(beam.PTransform):
...
>> beam.WindowInto(
window.FixedWindows(self.window_size))
pipeline
| "Read PubSub Messages"
>> beam.io.ReadFromPubSub(topic=input_topic)
| "Window into"
>> GroupWindowsIntoBatches(window_size)
Kafka IO
Java
PCollectoin<KV<String, String>> records =
pipeline
.apply("Rad From Kafka",
KafkaIO.<String, String>read()
.withConsumerConfigUpdates(ImmutableMap.of(
CosumerConfig
.AUTO_OFFSET_RESET_CONFIG, "earlies"))
.withBootstrapServers(options.getBootstrapServers())
.withTopics(<...list...>)
.withKeyDeserializersAndCoder(...))
.withValueDeserializerAndCoder(...)
.withoutMetadata())
Python
pipeline
| ReadFromKafka(
consumer_config={
'bootstrap.servers':bootstrap_servers},
topics=[topic])
BigTable IO
Java
p.appy("filterd read",
BittableIO.read()
.withProjectId(projectId)
.withInstanceId(instanceId)
.withTableId("table")
.withRowFilter(filter));
Reading with Prefix Scan
ByteKeyRange keyRange = ...;
p.appy("read",
BittableIO.read()
.withProjectId(projecctId)
.withInstanceId(instanceId)
.withTableId("table")
.withKeyRange(keyRange));
BigTable IO writing with additional actions
PCollection<KV<..., Iterable<Mutation>>> data = ...;
PCollection<BigtableWriteResult> writeResults =
data.apply("write",BittableIO.write()
.withProjectId("project")
.withInstanceId("instance")
.withTableId("table"))
.withWriteResults();
PCollection<...> moreData = ...;
moreData
.apply("wait for writes", Wait.on(writeResults))
.apply("do something", ParDo.of(...))
Avro IO
Java
PCollection<AvroAutoGenClass> records =
p.apply(AvroIO.read(AvroAutoGenClass.class)
.from("gs:...*.avro"));
Schema schema = new Schema.Parser()
.parse(new File("schema.avsc"));
PCollecction<GenericRecord> records =
p.apply(AvroIO.readGenericRecords(schema)
.from("gs:...-*.avro"));
Python
with beam.Pipeline() as p:
records = p | 'Read' >> beam.io.ReadFromAvro('/mypath/myavrofiles*')
Splittable DoFn
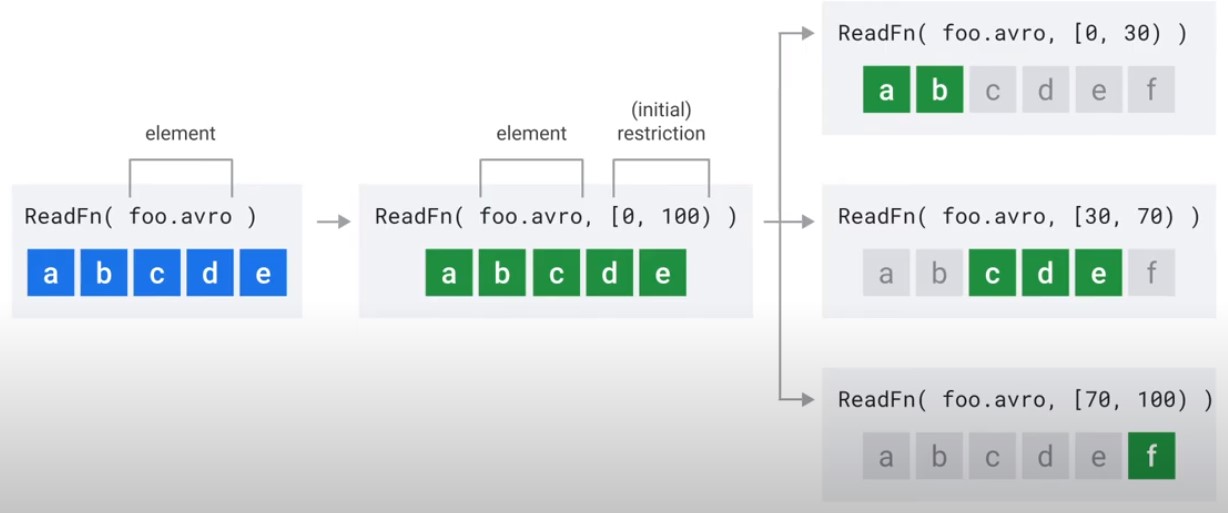
Java
@BoundPerElement
private static class FileToWordsFn extends DoFn<String,Integer> {
@GetInitialRestriction
public OffsetRange getInitialRestriction(
@Element String fileName) throws IOException {
return new OffsetRange(0,
new File(fileNam).length());
}
@ProcessElement
public void processElement(
@Element String fileName,
RestrictionTracker<OffsetRange, Long> tracker,
OutputReceiver<Integer> outputReceiver){...}
Python
class FileToWordsRestrictionProvider(
beam.io.RestrictionProvider):
def initial_restriction(self, file_name):
return OffsetRange(0,os.stat(file_name).st_size)
def create_tracker(self,restriction):
return beam.io.restriction_trackers.OffsetRestrictionTracker()
clas FileToWordsFn(beam.DoFn):
def process(...)
Beam Schemas
Convert Elements into Objects
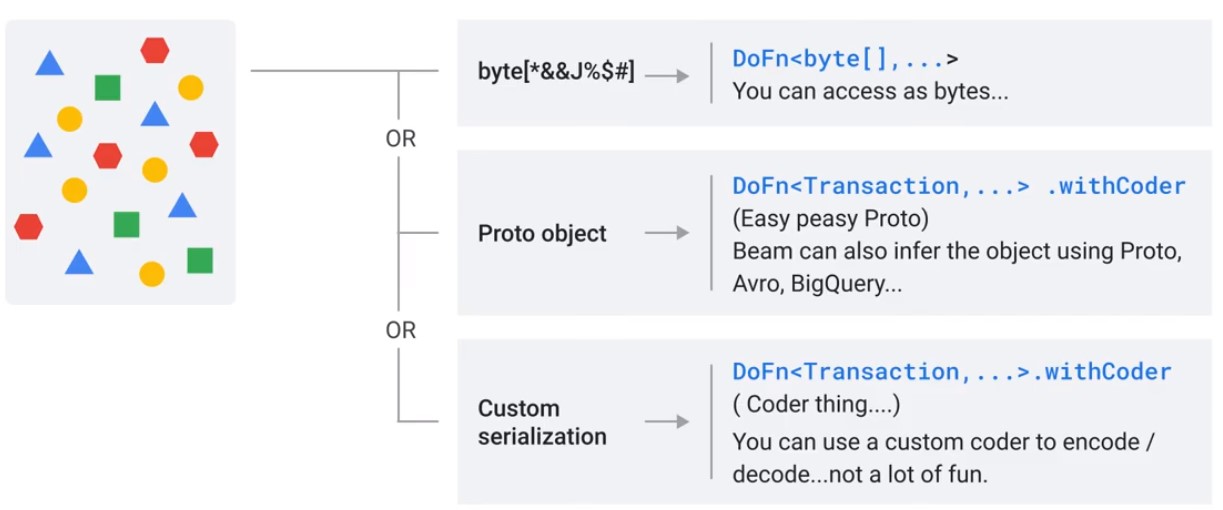
Schemas
- Describes a type in Terms of fields and values
- String names or numerical indexed
- Primitive Types int, long, string
- Some field can be marked optional
- Schemas can be nested
Code Examples
Filter Purchases
Without Schemas
purchases.apply(Filter.by(purchase -> {
return purchase.location.lat < 40.720 && purchase.location.lat > 40.699
&& purchase.location.lon < -73.969 && purchase.locatoin.lon > -74.747}));
With Schemas
purchases.apply(
Filter.whereFieldName("location.lat", (double lat) -> lat < 40.720 && lat > 40.699)
.whereFieldName("lcoation.lon", (double lon) -> lon < -73.969 && lon > -74.747));
Total Purchases per Transaction
PCollection<UserPurchases> userSums =
purchases.apply(Join.innerJoin(transactions).using("transactionId"))
.apply(Select.fieldNames("lhs.userId","rhs.totalPurchase"))
.apply(Group.byField("userId").aggregateField(Sum.ofLongs(),"totalPurchase"));
State and Timers
- Domain-specific triggering
- Slowly changing dimensions
- Stream joins
- Fine-grained aggregation
- Per-key workflows
States
Types of State Variables
| Type | Strength | Dataflow |
|---|---|---|
| Value | Read/write any value | yes |
| Bag | Cheap append no ordering on read | yes |
| Combining | Associatie/cummulative compaction | yes |
| Map | Read/write just keys you specify | yes |
| Set | Membership checking | no |
Stateful ParDo
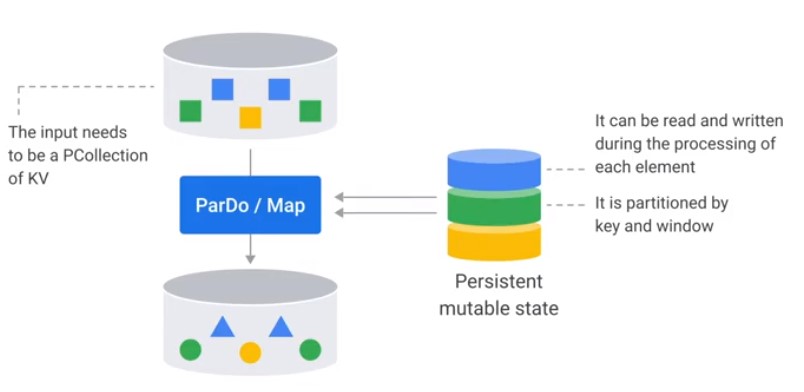
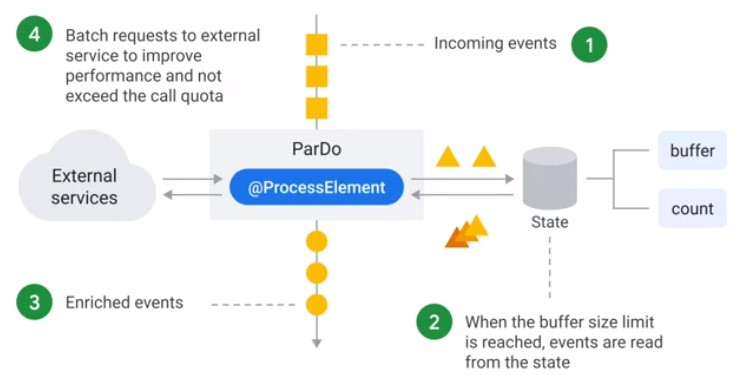
Accumulate Calls
class StatefulBufferingFn(beam.DoFn):
MAX_BUFFER_SIZE = 500;
BUFFER_STATE = BagStateSpec('buffer', EventCoder())
COUNT_STATE = CombiningValueStateSpec('count',VarIntCoder(),combiners.SumCombineFn())
def process(self, element,
buffer_state=beam.DoFn.StateParam(BUFFER_STATE),
count_state=beam.DoFn.StateParam(COUNT_STATE)):
buffer_state.add(element)
count_state.add(1)
count=count_state.read()
if count >= MAX_BUFFER_SIZE:
for event in buffer_state.read():
yield event
count_state.clear()
buffer_state.clear()
? What happens with the last Buffer, if it has not got enough messages to be cleared?
Timers

class StatefulBufferingFn(beam.DoFn):
...
EXPIRY_TIMER = TimerSpec('expiry', TimeDomatin.WATERMARK)
def process(self, element,
w = beam.DoFn.WindowParam,
...
expiry_timer=beam.DoFn.TimerParam(EXPIRY_TIMER)):
expiry_timer.set(w.end + ALLOWED_LATENESS)
@on_timer(EXPIRY_TIMER)
def_expiry(self,
buffer_state=beam.DoFn.StateParam(BUFFER_STATE),
count_state=beam.DoFn.StateParam(COUNT_STATE)):
events = buffer_state.read()
for event in events:
yield event
buffer_state.clear()
count_state.clear()
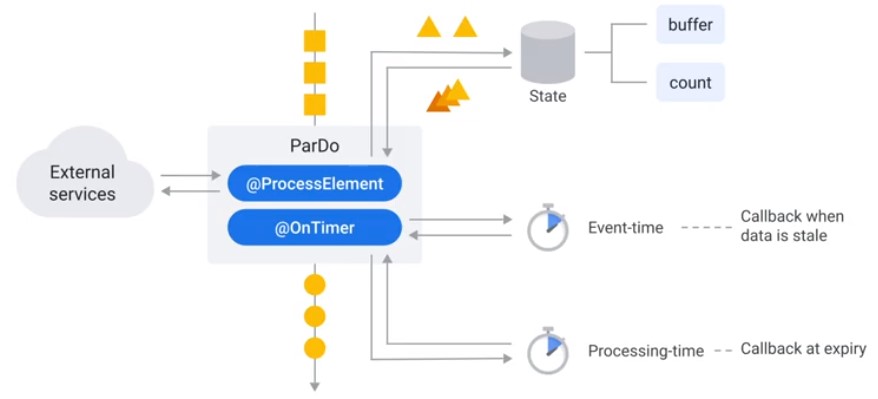
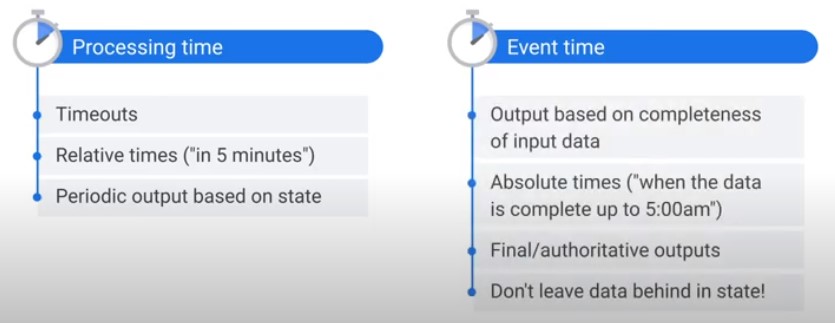
Best Practices
Handling Unprocessable Data
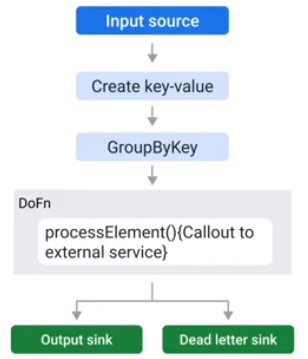
final TupleTag successTag;
final TupleTag deadLetterTag;
PCollection input = /* ... */;
PCollectionTuple outputTuple = input.apply(ParDo.of(new DoDn(){
@Override
void processElement(ProcessContext ctxt) {
try {
c.output(process(c.element));
} catch(MyException ex) {
//optional Logging at debug level
c.sideOutPut(deadLetterTag, c.element);
}
}
})).writeOutPutTags(successTag, TupleTagList.of(deadLetterTag));
// Write dead letter elements to separate sink
outputTuple.get(deadLetterTag).apply(BigQuery.write(...));
//Process the successful element differently
PCollection success = outputTuple.get(successTag);
Error Handling
- Errors are part of any data processing dataline
- Always wrap code in try-cat block
- In exception store exception to sink
AutoValue Code Generator
- Schemas are best way to represent objects in pipeline.
- There are still places where a POJO (Plain Old Java Objects) is needed.
- Use AutoValue class to generate POJOs.
Jandling JSON Data
PCollection<MyUserType> = json
.apply("Parse JSON to BEAM Rows", JsonToRow.withSchema(expectedSchema))
.apply("Convert to a user type with a compatible schema registered", Convet.to(MyUserType.class))
DoFn Lifecycle
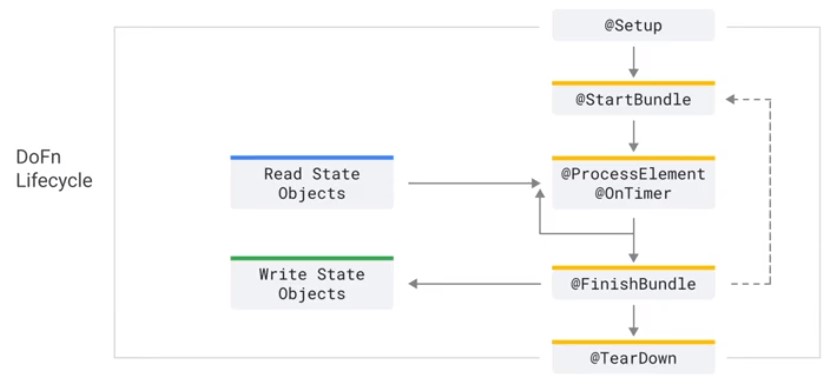
public class External extends DoFn{
@Override
public void startBundle(){
Instantiate your external service client (Static if threadsafe)
}
@Override
public void processElement(){
Call out to external service
}
@Override
puvlic void finishBundle(){
Shutdown your external service client if needed
}}
class MyDoFn(beam.DoFn):
def setup(self):
pass
def start_bundle(self):
pass
def process(self,element):
pass
def finish_bundle(self):
pass
def teardown(self):
pass
Pipeline Optimizations
- Filter data early
- Move any steps that reduce data volume up
- Apply data transformations serially to let Dataflow optimize DAG
- Transform applied serially are good candidates for graph optimization
- While working with external systems, look out for back pressure.
- Ensure external system are configured to handle peak volume.
SQL
Providing a schema enables SQL API.
| Input | Input | Input |
|---|---|---|
| BigQuery UI | Analytical queries over historical data | Data analyst |
| Dataflow SQL UI | Analytical queries over real-time data | Data analyst |
| Beam SQL | Integrating SQL within a Beam pipeline | Data engineer |
Beam SQL
- Works with stream and batch inputs
- Can be embedded in an existing pupeline usin SqlTransform
- Supports UDFs in Java
- Integrates with Schemas
- Stream aggregations support windows
Dialects
Apache Calcite
ZetaSQL
Dataflow SQL
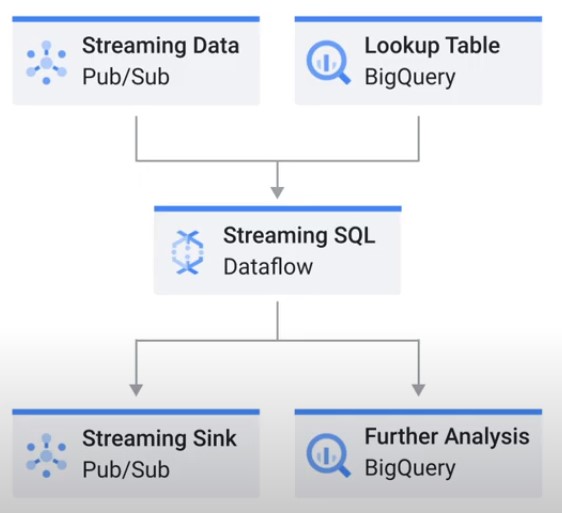
- Select from PubSub
- Join with batch data
- Aggregate over Window
-
$ gcloud dataflow sql query ‘select sum(foo) as baz, end_of_window from my_topic where something_is_true(bizzle) group by tumbling(timestamp, 1 hour)’
String sql1 = “select my_func(c1), c2 from pcollection”; PCollection
outpoutTableA = inputTableA.apply( SqlTransform .query(sql1) .addUdf("MY_FUNC", MY_FUNC.class, "FUNC");
Windowing in SQL
Fixed Windows
select
productId,
tumble_start("INTERVAL 10 SECOND") as period_start, count(transactionId) as num_purchases
from
pubsub.topic.`instant-insights`.`retaildemo-online-purchases-json` as pr
group by
productId,
tumble(pr.event_timestamp, "INTERVAL 10 SECOND");
Sliding Windows
select
productId,
hop_start("INTERVAL 10 SECOND","INTERVAL 30 SECOND") as period_start,
hop_end("INTERVAL 10 SECOND","INTERVAL 30 SECOND") as period_end,
count(transactionId) as num_purchases
from
pubsub.topic.`instant-insights`.`retaildemo-online-purchases-json` as pr
group by
productId,
hop(pr.event_timestamp,"INTERVAL 10 SECOND","INTERVAL 30 SECOND");
Session Windows
select
userId,
session_start("INTERVAL 10 MINUTE") as interval_start,
session_end("INTERVAL 10 MINUTE") as interval_end,
count(transactionId) as num_transactions
from
pubsub.topic.`instant-insights`.`retaildemo-online-purchases-json` as pr
group by
userId,
session(pr.event_timestamp, "INTERVAL 10 MINUTE");
Beam DataFrames
- Compatible with Pandas DataFrames
- Prallel processing with Beam model
- Provides access to familiar interfase within Beam pipeline
DataFrames GroupBy
pc = p | beam.Creaate(['strawberry','raspberry','blackberry','blueberry','bananan']
pc | GroupBy(lambda name: name[0])
DataFrames Transform
def my_function(df):
df['C'] = df.A + 2*df.B
result = df.groupby('C').sum().filter('A < 0')
return result
output = input | DataframeTransform(my_function)
Convert PCollection to Beam DataFrames
with beam.Pipeline() as p:
pc1 = ...
pc2 = ...
df1 = to_dataframe(pc1)
df2 = to_dataframe(pc2)
...
result = ...
result_pc = to_pcollection(result)
result_pc | beam.WriteToText(...)
Differences from standard Pandas
- Operations are deferred
- Result columns must be computable without access to the data
- PCollections in Beam are inherently unordered
Use case: Count Words
words = (
lines
| 'Split' >> beam.FlatMap(
lamda line: re.findall(r'[\w]+', line)).with_output_types(str)
| 'ToTows' >> beam.Map(lambda word: beam.Row(word=word)))
df = to_dataframe(words)
df['count'] = 1
counted = df.groupby['word'].sum()
counted.to_csv(known_args.output)
counted_pc = to_pcollection(counted, include_indexes=True)
_ = (
counted_pc
| beam.Filter(lambda row: row.count > 50)
| beam.Map(lambda row: f'{row.word}:{row.count}')
| beam.Map(print))
Beam Notebooks
gcp > Datflow > Notebooks
Add a transform
words = p | "read" >> beam.io.ReadFromPubSub(topic=topic)
windowed_words = (words
| "window" >> beam.WindowInto(beam.window.FixedWindows(10)))
windowed_words_counts = (windowed_words
| "count" >> beam.combiners.Count.PerElement())
Interactivity Optins before we run the Cell
Development to Production
from apache_beam.runners import DataflowRunner
options = pipeline_options.PipelineOptions()
runner = DataflowRunner()
runner.run_pipeline(p,options=options)
Cloud Monitoring
Job Information
Filtered view is saved in URL => Copy and Paste
- List
- Info
- Graph
- Metrics
Troubleshooting
Types of Troubles
- Failure in building the pipeline
- Validate “Beam Model” aspects
- Validate input/output specifications
- Reproduce it with direct runner, in a unit test
- Failure in starting the pipeline
- Failure during pipeline execution
- Performance problems
Performance
Topology
Filter early in the pipeline:
Read > Filter > Window > GBK
Coders
Econding and decoding cause larger overheads.
Windows
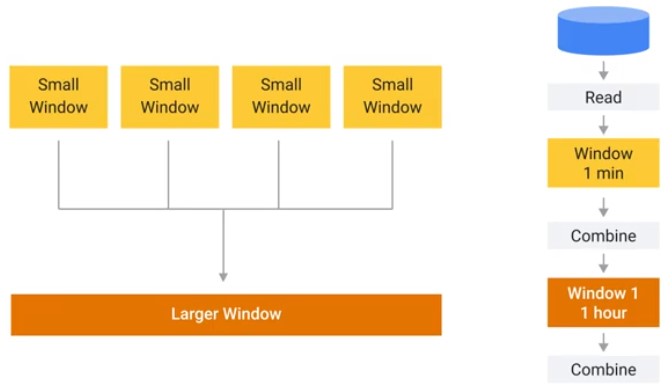
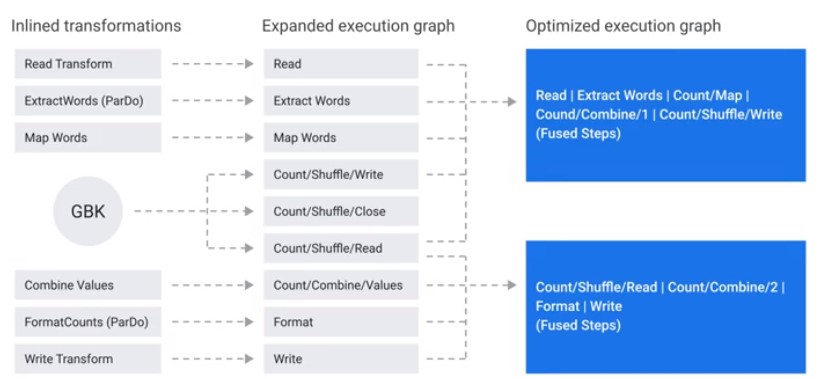
Graph Optimizations
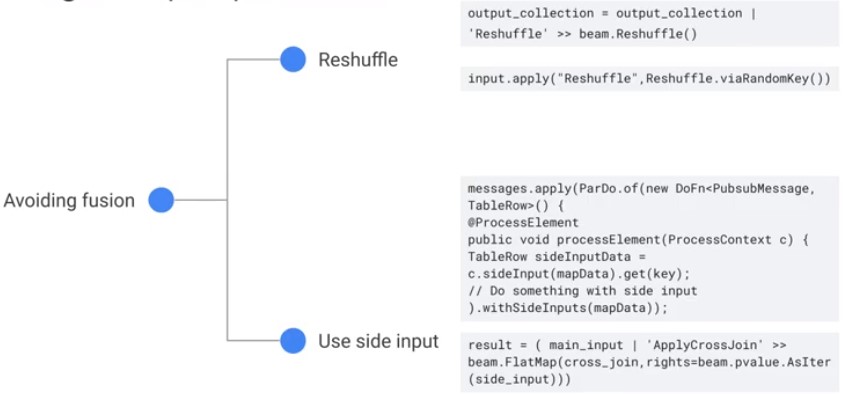
Fanout transformations: A single element outputs multiple elements. Avoid fusion, use side input.
Logging
- Balance between excessive logging vs no logging
- Avoid logging at info level agains PCollection element granularity
- Use a dead letter pattern followed by a count per window for reporting data errors.
Data Shape
Hot Key mitigation
Let Dataflow service log hotkeys with “hotKeyLoggingEnabled”
Key space and parallelism
- Low parallelism (to few keys)
- Increase number of keys
- Read from files, prefer splittable compression formats like Avro
- Too high parallelism
- If key space is large, consider unsing hashes separating keys
- Re-use processing keys from the past that are not active
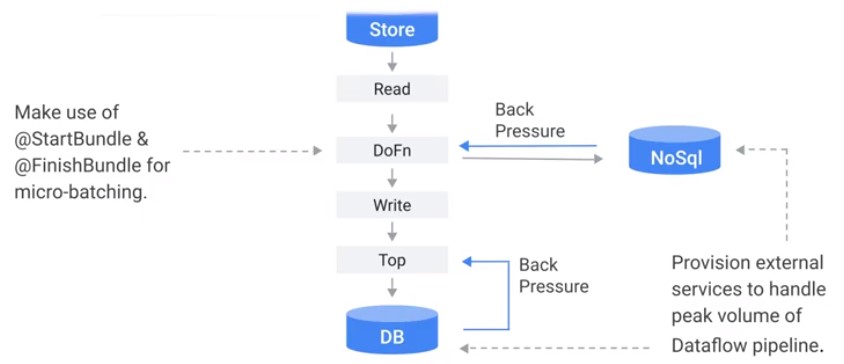
Source, Sinks & External Systems
- TextOP + Compressed file
Switch to: AcroIO, TextIO + Uncompressed
External System
Dataflow Shuffle
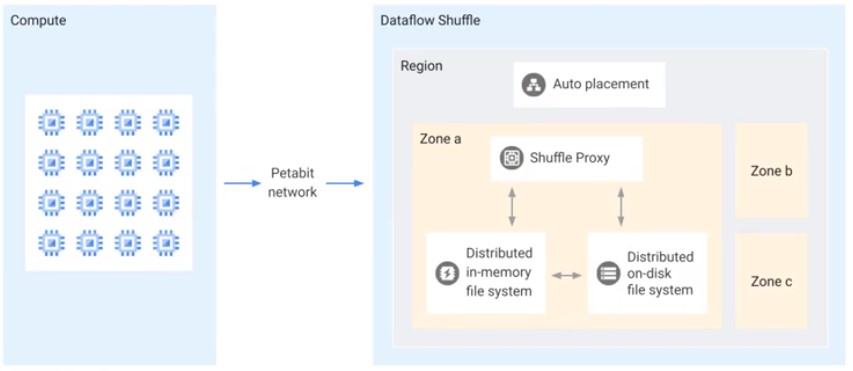
- Faste execution of batch pipeline
- Reduction of worker resources
- Better autoscaling
- Better fault tolerance
Testing and CI/CD Overview
Concepts
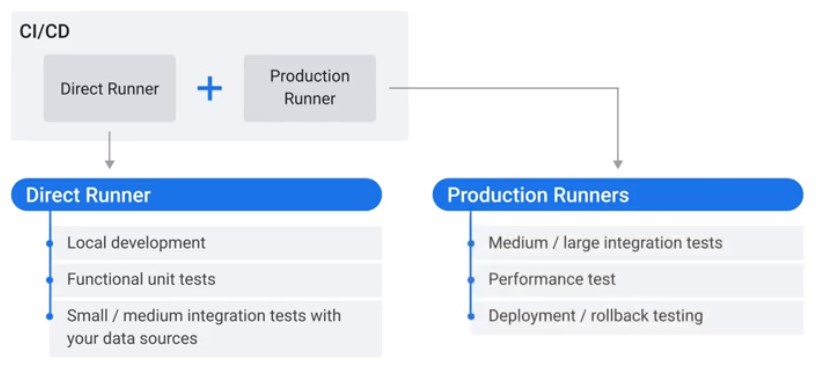
Direct Runner vs Production Runner
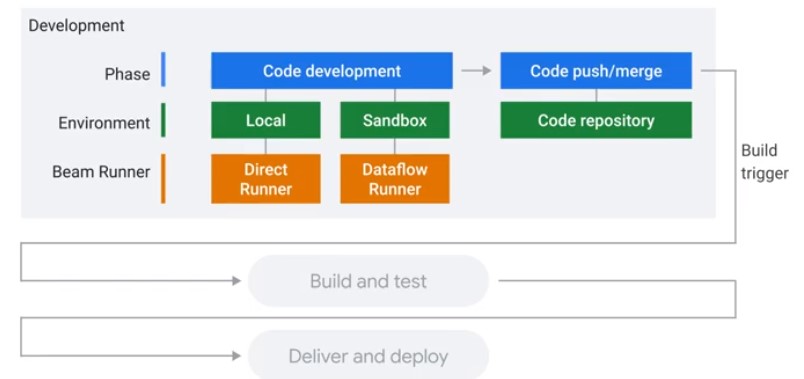
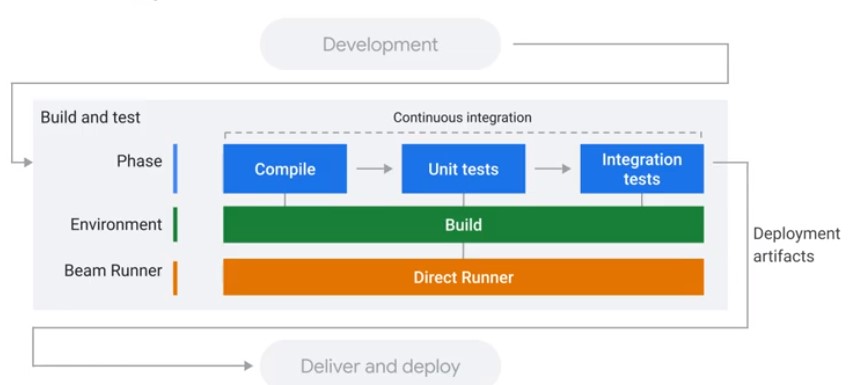
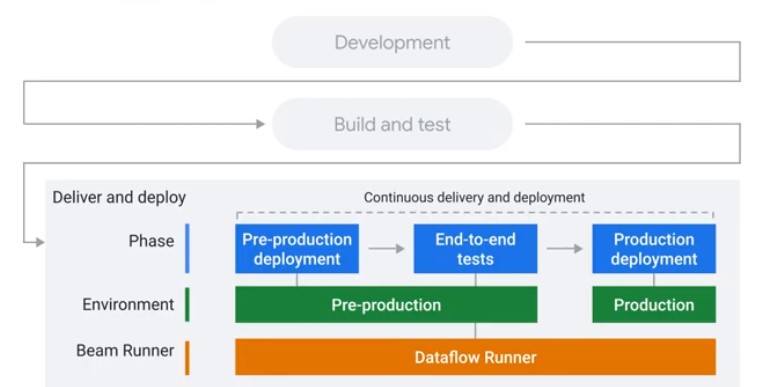
Unit Testing
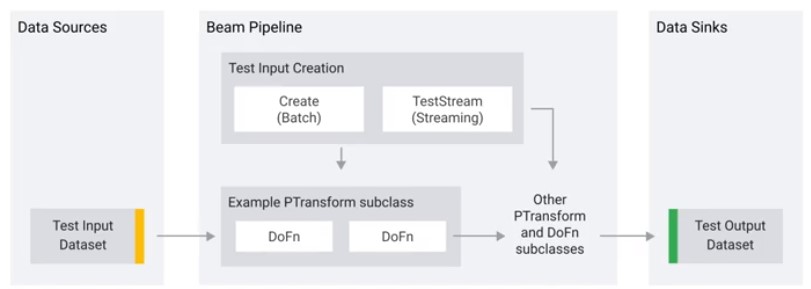
- Unit test in Beam are mini-pipelines that assert that the behaviour of small portions of out pipeline are correct.
- Unit test your DoFns or PTransforms
- Unit tests should run quickly, locally, and without dependencies on external systems.
-
JUnit 4 for unit testing
@Rule public TestPipeline p = TestPipeline.create();
@Test public void testASingleTransform() { // Setup your PCollection // from an in-memory or local data source. … // Apply your transform. PCollection
words = lines.apply(ParDo.of(new WordCount.ExtractWordsFn())); // Setup assertions on the pipeline. ... p.run(); }
Testing Classes
Do not use Anonymous DoFns. Prefer named Subclasses.
Java
@Rule
public final transient TestPipeline p = TestPipeline.create();
@Test
@Catergory(NeedsRunner.class)
public void myPipelineTest() throws Expectation {
final PCollection<String> pcol = d.apply(...)
PAssert.that(pcol).containsInAnyOrder(...);
p.run();
}
Python
with TestPipeline as P:
...
from apache_beam.testing.util import assert_that
from apache_beam.testing.util import equal_to
output = ...
# Check whether a PCollection
# contains some elements in any order.
assert_that(
output,
equal_to(["elem1","elem3","elem2"]))
Testing Windowing Behaviour
@Test
@Category(NeedsRunner.class)
public void testWindowedData() {
PCollection<String> input =
p.apply(Create.timestamped(
TimestampedValue.of("a", new Instant(0L)),
TimestampedValue.of("b", new Instant(0L)),
TimestampedValue.of("c", new Instant(0L)),
TimestampedValue.of("c", new instant(0L))
.plus(WINDOW_DURATION))
.withCoder(StringUtf8Coder.of()));
PCollection<KV<Sting, long>> windowedCount = input.apply(
Window.into(FixedWindows.of(WINDOW_DURATION))).apply(Count.perElement());
PAssert.that(windowedCount).containtsInAnyOrderd(
// Ouput from first window
KV.of("a", 2L),
KV.of("b", 1L),
KV.of("c", 1L),
KV.of("c", 1L));
p.run();
}
Test Streaming Pipelines
TestStream is a testing input that:
- Generates unbounded PCollectoin of elements
- Advances the watermar
- Processes time as elements are emitted
-
Stops producting output after all specified elements are emitted
@Test @Category(NeedsRunner.class) public void testDroppedLateData() { TestStream
input = TestStream.create(StringUtf8Coder.of()) .addElements( TimestampedValue.of("a", new Instant(0L)), TimestampedValue.of("b", new Instant(0L)), TimestampedValue.of("c", new Instant(0L)), TimestampedValue.of("c", new instant(0L)) .plus(Duration.standardMinutes(3)))) .advanceWatermarkTo(new instant(0L).plus(WINDOW_DURATION) .plus(Duration.standardMinutes(1)))) .addElements(TimestampedValue.of("c", new Instant(0L))) .advanceWatermarkToInfinity(); PCollection<KV<String, Long>> windowedCount = ... .withCoder(StringUtf8Coder.of())); PAssert.that(windowedCount).containtsInAnyOrderd( // Ouput from first window KV.of("a", 2L), KV.of("b", 1L), KV.of("c", 1L)); p.run(); }
Integration Testing
Test complete pipeline without sources and sinks.
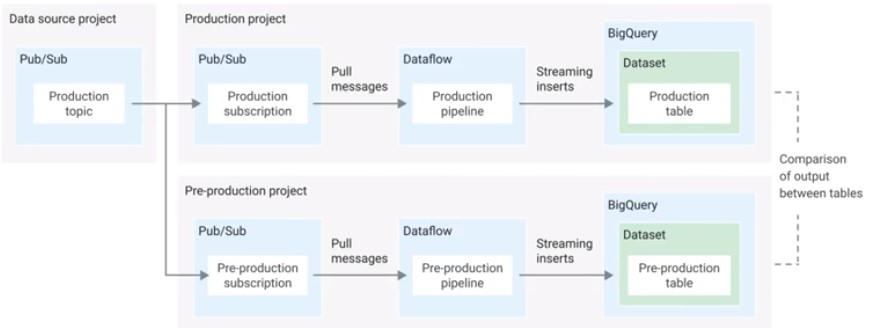
Batch Pipeline: Clone production pipeline into test-pipeline.
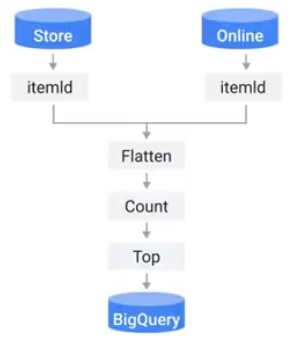
Streaming Pipeline: Create a new subscription to the topic.
Artifact Building
Use Beam 2.26 and higher
Deployment
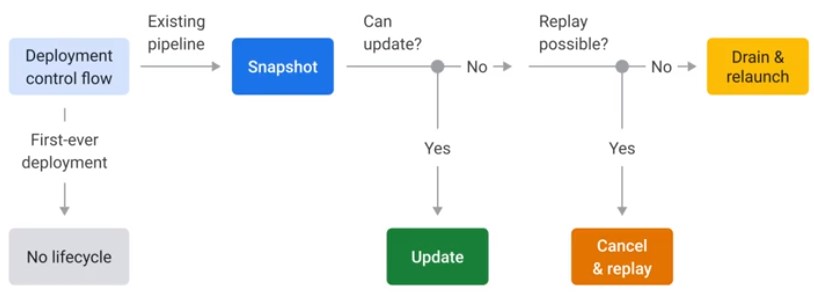
Deployment
- Direct launch: Launch from development environment
- Templates: Launch outside of developer environment
In-Flight
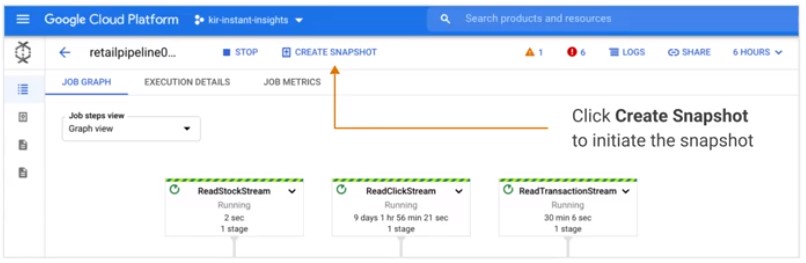
Snapshot
Save the state of streaming pipeline and launch new versions without losing state.
- Testing & Rollback updates
- Backup & Revocery
- Migrate to Streaming Engine
Create job from snapshot:
mvn -Pdataflow-runner compile exec:java \
-Dexec.mainClass=org.apache.beacm.examples.WordCount \
-Dexec.args="--project=PROJECT_ID \
--stagingLocation=gs://STORAGE_BUCKET/staging/ \
--inputFile=gs://apache-beam-samples/shakespeare/* \
--output=gs://STORAGE_BUCKET/output \
--runner=DataflowRunner \
--enableStreamingEngine \
--createFromSnapshot=SNAPSHOT_ID \
--region="REGION"
Update
Replace you existing job with a new job that runs updated pipeline code
- Improve pipeline code
- Fix bugs in pipeline code
- Update your pipeline
- Account for changes in data source
Java
mvn -Pdataflow-runner compile exec:java \
-Dexec.mainClass=org.apache.beam.examples.WordCound \
-Dexec.args="--project=PROJECT_ID \
--stagingLocations=gs://STORAGE_BUCKET/staging/ \
--inputFile=gs://apache-beam-samples/shakespeare/* \
--ouput=gs://STORAGE_BUCKET/ouput \
--runner=DataflowRunner \
--update \
--jobName [prior job name] \
--transformNameMapping='{"oldTransform1":"newTransform1","oldTransform2":"newTransform2",...}
--reagion=REGION"
Python
python -m apache_beam.examples.wordcount \
--project $PROJECT \
--staging_location gs://$BUCKET/tmp/
--input gs://dataflow-samples/shakespeare/kinglear.txt \
--output gs://$BUCKET/results/outputs \
--runner DataflowRunner \
--update \
--job_name [prior job name] \
--transform_name_mapping=='{oldTransform1":"newTransform1","oldTransform2":"newTransform2",...}'
--region $REGION \
Compatibility Failures
- No transform mapping
- Add/remoge side input
- Change coders
- Switching locations
- Removing stateful operations
Termination
- Drain: Stop data ingestion and continue data compute
- No data is lost
- Incomplete aggregations
- Cancel: Stop ingestion and compute immediately
Decission Tree
Reliability
- Batch
- Rerun
- Source Data not lost
- Stream
Failure Types
| User Code & Data Shape | Outages |
|---|---|
| Transient errors | Service outage |
| Corrupted data | Zonal outage |
| Regional outage |
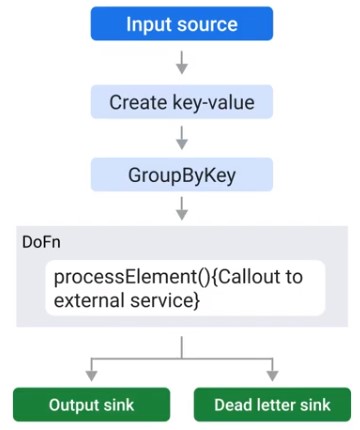
Dead Letter sink
final TupleTag successTag;
final TupleTag deadLetterTag;
PCollection input = /* ... */
PCollection Tuple ouputTuple = input.apply(ParDo.of(new DoFn() {
@Override
void processElement(ProcessContext ctxt) {
try {
c.output(process(c.element));
} catch(MyException ex) {
// Optional Loccing at debug level
c.sideOutPut(deadLetterTag, c.element);
}
})).writeOutPutTags(successTag, TupleTagList.of(deadLetterTag));
// Write dead letter elements to separate sink
outputTuple.get(deadLetterTag).apply(BigQuery.write(...));
// Process the successful element differently.
PCollection success = outputTuple.get(successTag);
Monitoring and Alerting
Streaming jobs try to re-run indefinetly.
- Catch issues before they bring down production systems.
- Datflow Job Metrics tab provides an integrated monitoring experiencce
- Cloud Monitoring integration extends capabilities
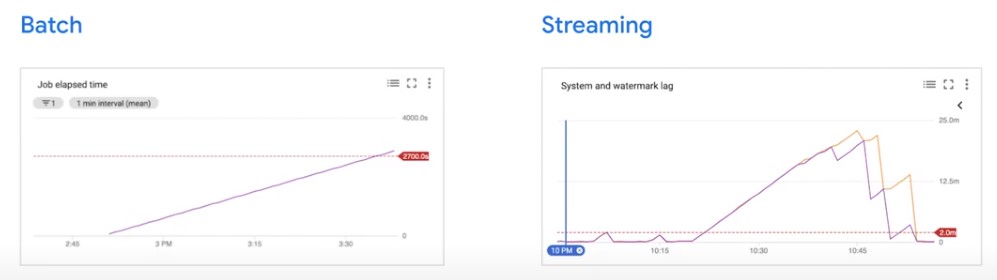
Batch
Streaming
Geolocation
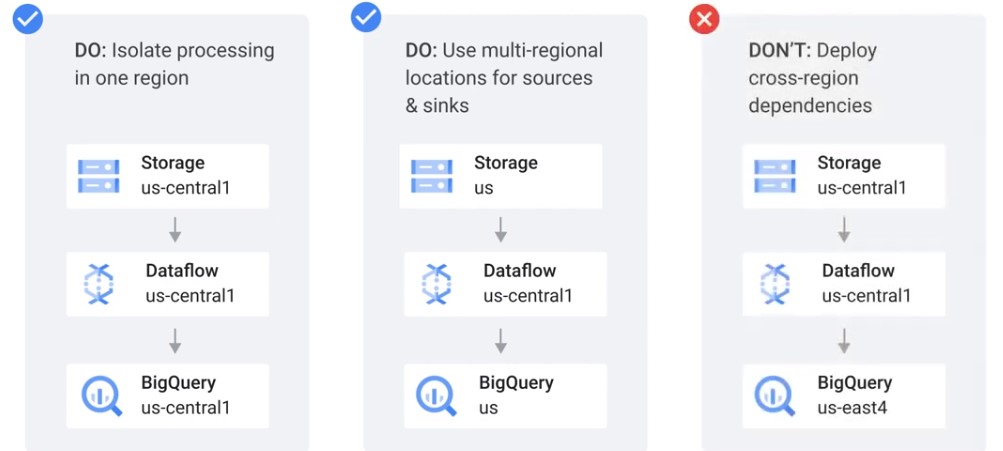
Disaster Recovery
Pub/Sub
Make Snapshot
gcloud pubsub snapshots create my-snapshot --subscription=my-sub
Stop an drain Pipeline
gcloud dataflow jobs drain [job-id]
Seek your subscription to the snapshot
gcloud pubsub subscriptions seek my-sub --snapshot=my-snapshot
Resubmit pipeline
gcloud dataflow jobs run my-job-name --gcs_locatoin=my_gcs_bucket
Dataflow
- Restart pipeline without processing in-flight data
- No data loss with minimal downtime
- Option to create a Snapshot with Pub/Sub source
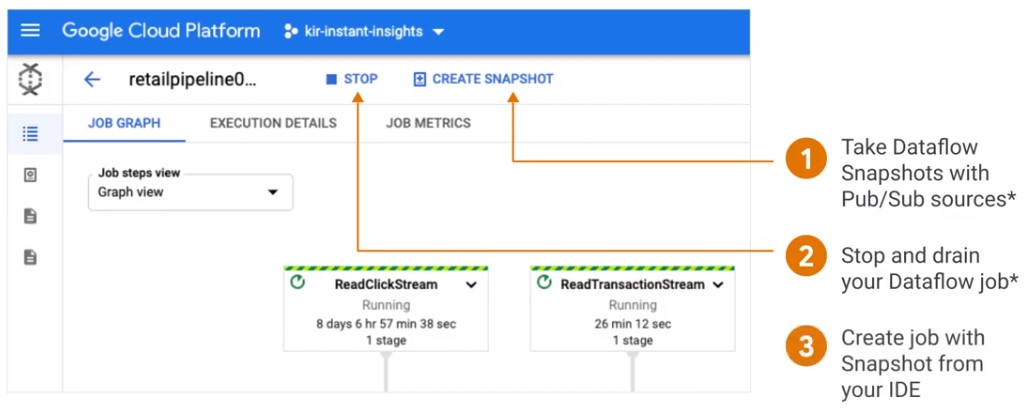
- Schedule Weely Dataflow Snapshot with CloudComposer
- Snapshots are stored in the region of their job.
High Availability
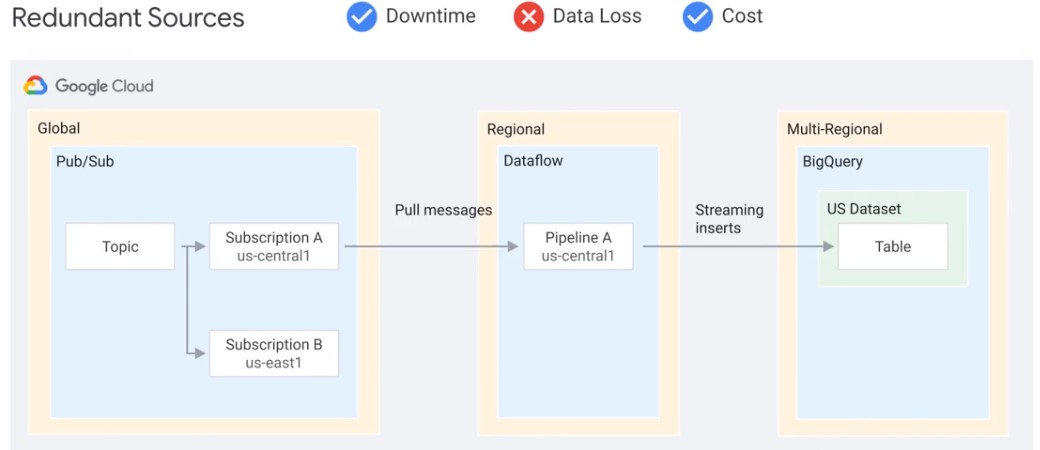
Redundand Sources
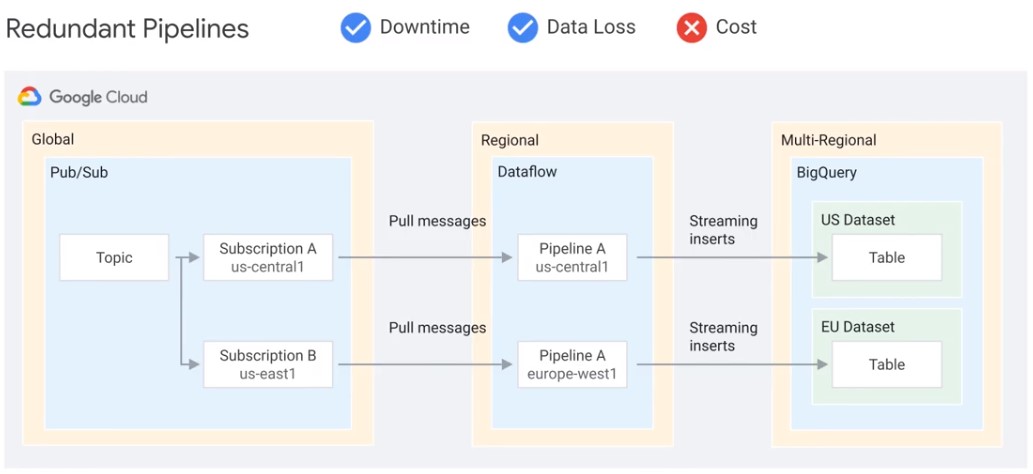
Redundand Pipelines
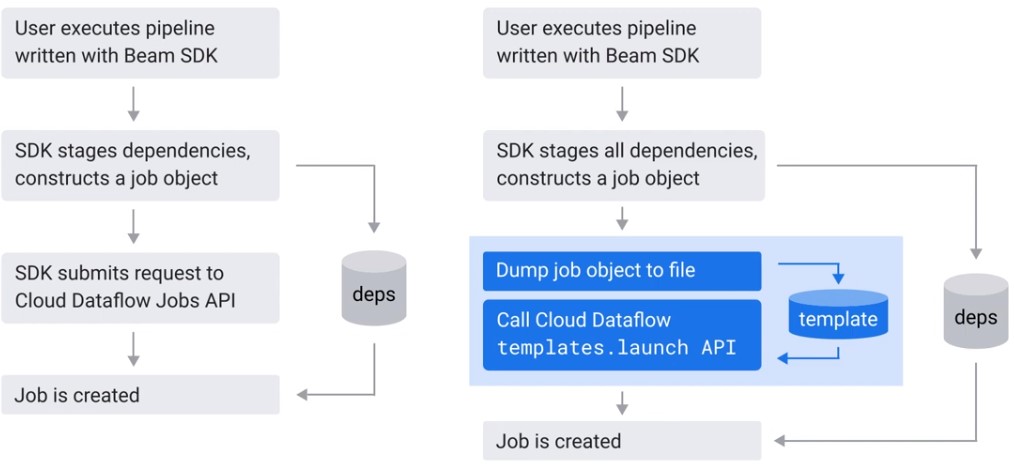
Flex Templates
Classic Templates
Flex Templates
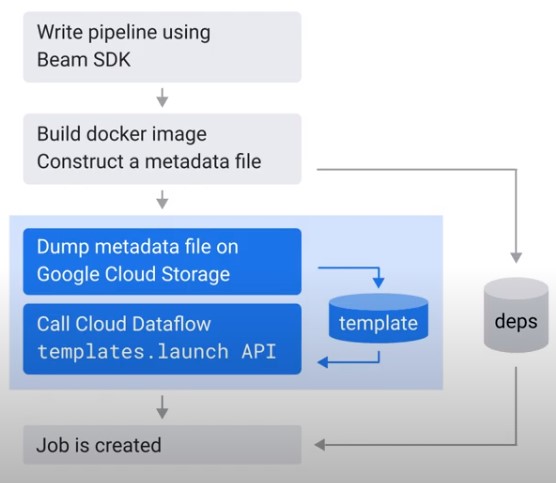

Create
metadata.json
{
"name":"PubSub To Biggquery",
"description":"An Apache Beam streaming pipeline that reads JSON",
"parameters":[
{
"name":"inputSubscription",
"label":"Pub/Sub input subscription"
"helpText":"Pub7Sub subscription to read from",
"regexes":["[a-zA-Z][-_.~+%]"]
},
{
"name":"outputTable",
"label":"BigQuery outputTable",
"helpText":"Write to table",
"regexes":["^:"]
}
]
}
Build the flex template
$ gcloud dataflow flex-template build "$TEMPLATE_SPEC_PATH" \
--image-gcr-path "$TEMPLATE_IMAGE" \
--sdk-language "JAVA" \
--flex-template-base-image JAVA8 \
--metadata-file "metadata.json" \
--jar "target/pubsub-bigquery-1.0.jar" \
--env FLEXTEMPLATE_JAVA_MAIN_CLASS="com.google.cloud.PubSuibBigquery"
Launch Flex Template
Console
$ gcloud dataflow flex-template run "job-name-`date +%Y%m%d-%H-%M-%S`" \
--template-file-gcs-location "$TEMPLATE_PATH" \
--parameters inputSubscription="$SUBSCRIPTION" \
--parameters outputTable="$PROJECT:$DATASET.$TABLE" \
--region "$REGION"
REST API
$ curl -X POST \
"https://dataflow.googleapis.com/v1b3/projects/$PROJECT/locations/${REGION}/flexTemplates:launch" \
-H "Content-Type: application/json" \
-H "Authorization: Bearer $(gcloud auth print-access-token)" \
-d '{
"launch_parameter":{
"jobName":"job-name-`date +%Y%m%d-%H%M%S`",
"parameters":{
"inputSubscription":"'SUBSCRIPTION'",
"outputTable":"'$PROJECT:$DATASET.$TABLE'"
},
"containerSpecGcsPath":"'$TEMPLATE_PATH'"
}
}'
Cloud Scheduler
$ gcloud scheduler jobs create http scheduler-job --schedule="*/30 * * * *"
--uri="https://dataflow.googleapis.com/v1b3/projects/$PROJECT/locations/${REGION}/flexTemplates:launch" --http-method=POST \
--oauth-service-account-email=email@project.iam.gserviceaccount.com \
--message-body=' {
"launch_parameter":{
"jobName":"job-name"
"parameters":{
"inputSubscription":"'$SUBSCRIPTION'",
"outputTable":"'$PROJECT:$DATASTE.$TABLE'"
},
"containerSpecGcsPath":"'$TEMPLATE_PATH'"
}
}'
